Genome-Wide Analysis of Multidrug and Toxic Compound Extrusion (MATE) Family in Gossypium raimondii and Gossypium arboreum and Its Expression Analysis Under Salt, Cadmium, and Drought Stress
- PMID: 29794162
- PMCID: PMC6027885
- DOI: 10.1534/g3.118.200232
Genome-Wide Analysis of Multidrug and Toxic Compound Extrusion (MATE) Family in Gossypium raimondii and Gossypium arboreum and Its Expression Analysis Under Salt, Cadmium, and Drought Stress
Abstract
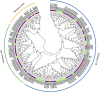
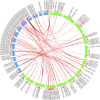
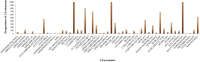
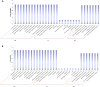
The extrusion of toxins and substances at a cellular level is a vital life process in plants under abiotic stress. The multidrug and toxic compound extrusion (MATE) gene family plays a large role in the exportation of toxins and other substrates. We carried out a genome-wide analysis of MATE gene families in Gossypium raimondii and Gossypium arboreum and assessed their expression levels under salt, cadmium and drought stresses. We identified 70 and 68 MATE genes in G. raimondii and G. arboreum, respectively. The majority of the genes were predicted to be localized within the plasma membrane, with some distributed in other cell parts. Based on phylogenetic analysis, the genes were subdivided into three subfamilies, designated as M1, M2 and M3. Closely related members shared similar gene structures, and thus were highly conserved in nature and have mainly evolved through purifying selection. The genes were distributed in all chromosomes. Twenty-nine gene duplication events were detected, with segmental being the dominant type. GO annotation revealed a link to salt, drought and cadmium stresses. The genes exhibited differential expression, with GrMATE18, GrMATE34, GaMATE41 and GaMATE51 significantly upregulated under drought, salt and cadmium stress, and these could possibly be the candidate genes. Our results provide the first data on the genome-wide and functional characterization of MATE genes in diploid cotton, and are important for breeders of more stress-tolerant cotton genotypes.
Keywords: GO annotation; GenPred; Genomic Selection; Gossypium arboreum; Gossypium raimondii; MATE genes; Shared Data Resources; cadmium; drought; phylogenetic tree analysis; stress.
Copyright © 2018 Lu et al.
Figures



Similar articles
-
Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants.Int J Mol Sci. 2018 Sep 5;19(9):2625. doi: 10.3390/ijms19092625. Int J Mol Sci. 2018. PMID: 30189594 Free PMC article.
-
Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton.BMC Genet. 2019 Jul 23;20(1):62. doi: 10.1186/s12863-019-0756-6. BMC Genet. 2019. PMID: 31337336 Free PMC article.
-
Genome-wide characterization and expression analysis of the aldehyde dehydrogenase (ALDH) gene superfamily under abiotic stresses in cotton.Gene. 2017 Sep 10;628:230-245. doi: 10.1016/j.gene.2017.07.034. Epub 2017 Jul 12. Gene. 2017. PMID: 28711668
-
Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton.Int J Biol Macromol. 2021 Dec 1;192:1058-1074. doi: 10.1016/j.ijbiomac.2021.10.038. Epub 2021 Oct 14. Int J Biol Macromol. 2021. PMID: 34656543 Review.
-
Identification of BR biosynthesis genes in cotton reveals that GhCPD-3 restores BR biosynthesis and mediates plant growth and development.Planta. 2021 Sep 17;254(4):75. doi: 10.1007/s00425-021-03727-9. Planta. 2021. PMID: 34533620 Review.
Cited by
-
Structural and Functional Characterization at the Molecular Level of the MATE Gene Family in Wheat in Silico.Contrast Media Mol Imaging. 2022 Aug 29;2022:9289007. doi: 10.1155/2022/9289007. eCollection 2022. Contrast Media Mol Imaging. 2022. PMID: 39281829 Free PMC article.
-
Genome-wide characterization of MATE gene family and expression profiles in response to abiotic stresses in rice (Oryza sativa).BMC Ecol Evol. 2021 Jul 9;21(1):141. doi: 10.1186/s12862-021-01873-y. BMC Ecol Evol. 2021. PMID: 34243710 Free PMC article.
-
Genome-Wide Identification, Expression Analysis under Abiotic Stress and Co-Expression Analysis of MATE Gene Family in Torreya grandis.Int J Mol Sci. 2024 Mar 29;25(7):3859. doi: 10.3390/ijms25073859. Int J Mol Sci. 2024. PMID: 38612669 Free PMC article.
-
Molecular and biochemical components associated with chilling tolerance in tomato: comparison of different developmental stages.Mol Hortic. 2024 Sep 5;4(1):31. doi: 10.1186/s43897-024-00108-0. Mol Hortic. 2024. PMID: 39232835 Free PMC article.
-
Genome-wide identification, characterization and expression profiles of FORMIN gene family in cotton (Gossypium Raimondii L.).BMC Genom Data. 2024 Dec 18;25(1):105. doi: 10.1186/s12863-024-01285-z. BMC Genom Data. 2024. PMID: 39695391 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
