A 2-transcript host cell signature distinguishes viral from bacterial diarrhea and it is influenced by the severity of symptoms
- PMID: 29795312
- PMCID: PMC5966427
- DOI: 10.1038/s41598-018-26239-1
A 2-transcript host cell signature distinguishes viral from bacterial diarrhea and it is influenced by the severity of symptoms
Abstract
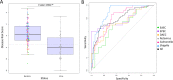
Recently, a biomarker signature consisting of 2-transcript host RNAs was proposed for discriminating bacterial from viral infections in febrile children. We evaluated the performance of this signature in a different disease scenario, namely a cohort of Mexican children (n = 174) suffering from acute diarrhea of different infectious etiologies. We first examined the admixed background of the patients, indicating that most of them have a predominantly Native American genetic ancestry with a variable amount of European background (ranging from 0% to 57%). The results confirm that the RNA test can discriminate between viral and bacterial causes of infection (t-test; P-value = 6.94×10-11; AUC = 80%; sensitivity: 68% [95% CI: 55%-79%]; specificity: 84% [95% CI: 78%-90%]), but the strength of the signal differs substantially depending on the causal pathogen, with the stronger signal being that of Shigella (P-value = 3.14 × 10-12; AUC = 89; sensitivity: 70% [95% CI: 57%-83%]; specificity: 100% [95% CI: 100%-100%]). The accuracy of this test improves significantly when excluding mild cases (P-value = 2.13 × 10-6; AUC = 85%; sensitivity: 79% [95% CI: 58%-95%]; specificity: 78% [95% CI: 65%-88%]). The results broaden the scope of previous studies by incorporating different pathogens, variable levels of disease severity, and different ancestral background of patients, and add confirmatory support to the clinical utility of these 2-transcript biomarkers.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

