NOTCH3 inactivation increases triple negative breast cancer sensitivity to gefitinib by promoting EGFR tyrosine dephosphorylation and its intracellular arrest
- PMID: 29795369
- PMCID: PMC5968025
- DOI: 10.1038/s41389-018-0051-9
NOTCH3 inactivation increases triple negative breast cancer sensitivity to gefitinib by promoting EGFR tyrosine dephosphorylation and its intracellular arrest
Abstract
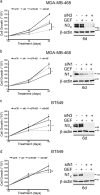
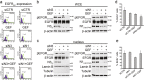
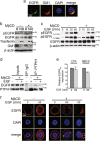
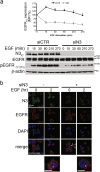
Notch dysregulation has been implicated in numerous tumors, including triple-negative breast cancer (TNBC), which is the breast cancer subtype with the worst clinical outcome. However, the importance of individual receptors in TNBC and their specific mechanism of action remain to be elucidated, even if recent findings suggested a specific role of activated-Notch3 in a subset of TNBCs. Epidermal growth factor receptor (EGFR) is overexpressed in TNBCs but the use of anti-EGFR agents (including tyrosine kinase inhibitors, TKIs) has not been approved for the treatment of these patients, as clinical trials have shown disappointing results. Resistance to EGFR blockers is commonly reported. Here we show that Notch3-specific inhibition increases TNBC sensitivity to the TKI-gefitinib in TNBC-resistant cells. Mechanistically, we demonstrate that Notch3 is able to regulate the activated EGFR membrane localization into lipid rafts microdomains, as Notch3 inhibition, such as rafts depletion, induces the EGFR internalization and its intracellular arrest, without involving receptor degradation. Interestingly, these events are associated with the EGFR tyrosine dephosphorylation at Y1173 residue (but not at Y1068) by the protein tyrosine phosphatase H1 (PTPH1), thus suggesting its possible involvement in the observed Notch3-dependent TNBC sensitivity response to gefitinib. Consistent with this notion, a nuclear localization defect of phospho-EGFR is observed after combined blockade of EGFR and Notch3, which results in a decreased TNBC cell survival. Notably, we observed a significant correlation between EGFR and NOTCH3 expression levels by in silico gene expression and immunohistochemical analysis of human TNBC primary samples. Our findings strongly suggest that combined therapies of TKI-gefitinib with Notch3-specific suppression may be exploited as a drug combination advantage in TNBC treatment.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

