Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides
- PMID: 29795541
- PMCID: PMC6544545
- DOI: 10.1038/s41564-018-0164-0
Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides
Abstract
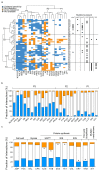
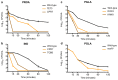
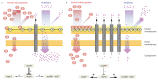
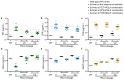
Antimicrobial peptides are promising alternative antimicrobial agents. However, little is known about whether resistance to small-molecule antibiotics leads to cross-resistance (decreased sensitivity) or collateral sensitivity (increased sensitivity) to antimicrobial peptides. We systematically addressed this question by studying the susceptibilities of a comprehensive set of 60 antibiotic-resistant Escherichia coli strains towards 24 antimicrobial peptides. Strikingly, antibiotic-resistant bacteria show a high frequency of collateral sensitivity to antimicrobial peptides, whereas cross-resistance is relatively rare. We identify clinically relevant multidrug-resistance mutations that increase bacterial sensitivity to antimicrobial peptides. Collateral sensitivity in multidrug-resistant bacteria arises partly through regulatory changes shaping the lipopolysaccharide composition of the bacterial outer membrane. These advances allow the identification of antimicrobial peptide-antibiotic combinations that enhance antibiotic activity against multidrug-resistant bacteria and slow down de novo evolution of resistance. In particular, when co-administered as an adjuvant, the antimicrobial peptide glycine-leucine-amide caused up to 30-fold decrease in the antibiotic resistance level of resistant bacteria. Our work provides guidelines for the development of efficient peptide-based therapies of antibiotic-resistant infections.
Conflict of interest statement
The authors declare no competing interests. BB and IN had consulting positions at SeqOmics Biotechnology Ltd. at the time the study was conceived. SeqOmics Biotechnology Ltd. was not directly involved in the design and execution of the experiments or in the writing of the manuscript. This does not alter the author’s adherence to all the Nature policies on sharing data and materials.
Figures


References
-
- Imamovic L, Sommer MO. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013;5:204ra132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

