An epigenetic classifier for early stage lung cancer
- PMID: 29796119
- PMCID: PMC5964676
- DOI: 10.1186/s13148-018-0502-3
An epigenetic classifier for early stage lung cancer
Abstract
Background: Methylated genes detected in sputum are promise biomarkers for lung cancer. Yet the current PCR technologies for quantification of DNA methylation and diagnostic value of the sputum biomarkers are not sufficient to be used for lung cancer early detection. The emerging droplet digital PCR (ddPCR) is a straightforward means for precise, direct, and absolute quantification of nucleic acids. Here, we investigate whether ddPCR can sensitively and robustly quantify DNA methylation in sputum for more precise diagnosis of lung cancer.
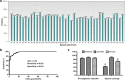
Results: First, the analytic performance of methylation-specific ddPCR (ddMSP) and quantitative methylation-specific PCR (qMSP) is determined in methylated and unmethylated DNA samples. Second, 29 genes, previously proposed as potential sputum biomarkers for lung cancer, are analyzed by using ddMSP in a training set of 127 lung cancer patients and 159 controls. ddMSP has higher sensitivity, precision, and reproducibility for quantification of methylation compared with qMSP (all p < 0.05). A classifier comprising four sputum methylation biomarkers for lung cancer is developed by using ddMSP, producing 86.6% sensitivity and 90.6% specificity, independent of stage and histology of lung cancer (all p > 0.05). The classifier has higher accuracy compared with sputum cytology (88.8 vs. 70.6%, p < 0.01). The diagnostic performance is confirmed in a testing set of 89 cases and 107 controls.
Conclusions: ddMSP is a robust tool for reliable quantification of DNA methylation in sputum, and the epigenetic classifier could help diagnose lung cancer at the early stage.
Keywords: DNA methylation; Diagnosis; Lung cancer; Sputum; ddPCR.
Conflict of interest statement
This study was supported in part by a grant for cancer research from the Jiangsu Province Hospital of Nanjing University of Chinese Medicine. The study was approved by the Nanjing University of Chinese Medicine Institutional Review Board and adhered to the Declaration of Helsinki, and all patients provided written informed consent.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum.Clin Cancer Res. 2017 Apr 15;23(8):1998-2005. doi: 10.1158/1078-0432.CCR-16-1371. Epub 2016 Oct 11. Clin Cancer Res. 2017. PMID: 27729459 Free PMC article.
-
Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection.Clin Epigenetics. 2016 Oct 19;8:109. doi: 10.1186/s13148-016-0275-5. eCollection 2016. Clin Epigenetics. 2016. PMID: 27777637 Free PMC article.
-
MethyLight droplet digital PCR for detection and absolute quantification of infrequently methylated alleles.Epigenetics. 2015;10(9):803-9. doi: 10.1080/15592294.2015.1068490. Epub 2015 Jul 17. Epigenetics. 2015. PMID: 26186366 Free PMC article.
-
Use of DNA methylation patterns for early detection and management of lung cancer: Are we there yet?Oncol Res. 2025 Mar 19;33(4):781-793. doi: 10.32604/or.2024.057231. eCollection 2025. Oncol Res. 2025. PMID: 40191732 Free PMC article. Review.
-
The Indirect Efficacy Comparison of DNA Methylation in Sputum for Early Screening and Auxiliary Detection of Lung Cancer: A Meta-Analysis.Int J Environ Res Public Health. 2017 Jun 23;14(7):679. doi: 10.3390/ijerph14070679. Int J Environ Res Public Health. 2017. PMID: 28644424 Free PMC article. Review.
Cited by
-
Cancer Detection and Classification by CpG Island Hypermethylation Signatures in Plasma Cell-Free DNA.Cancers (Basel). 2021 Nov 9;13(22):5611. doi: 10.3390/cancers13225611. Cancers (Basel). 2021. PMID: 34830765 Free PMC article.
-
Current and Emerging Applications of Droplet Digital PCR in Oncology: An Updated Review.Mol Diagn Ther. 2022 Jan;26(1):61-87. doi: 10.1007/s40291-021-00562-2. Epub 2021 Nov 13. Mol Diagn Ther. 2022. PMID: 34773243 Review.
-
Integrated analysis of miRNAs and DNA methylation identifies miR-132-3p as a tumor suppressor in lung adenocarcinoma.Thorac Cancer. 2020 Aug;11(8):2112-2124. doi: 10.1111/1759-7714.13497. Epub 2020 Jun 4. Thorac Cancer. 2020. PMID: 32500672 Free PMC article.
-
Precise diagnosis of three top cancers using dbGaP data.Sci Rep. 2021 Jan 12;11(1):823. doi: 10.1038/s41598-020-80832-x. Sci Rep. 2021. PMID: 33436913 Free PMC article.
-
Mechanism study of serum extracellular nano-vesicles miR-412-3p targeting regulation of TEAD1 in promoting malignant biological behavior of sub-centimeter lung nodules.Cancer Biomark. 2024;41(1):69-82. doi: 10.3233/CBM-240137. Cancer Biomark. 2024. PMID: 39269825 Free PMC article.
References
-
- Blumer G. Cigarette smoking and cancer of the lung. Ill Med J. 1951;100:98–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

