Demographic History and Genetic Adaptation in the Himalayan Region Inferred from Genome-Wide SNP Genotypes of 49 Populations
- PMID: 29796643
- PMCID: PMC6063301
- DOI: 10.1093/molbev/msy094
Demographic History and Genetic Adaptation in the Himalayan Region Inferred from Genome-Wide SNP Genotypes of 49 Populations
Abstract
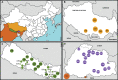
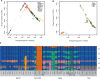
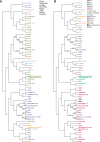
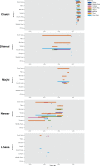
We genotyped 738 individuals belonging to 49 populations from Nepal, Bhutan, North India, or Tibet at over 500,000 SNPs, and analyzed the genotypes in the context of available worldwide population data in order to investigate the demographic history of the region and the genetic adaptations to the harsh environment. The Himalayan populations resembled other South and East Asians, but in addition displayed their own specific ancestral component and showed strong population structure and genetic drift. We also found evidence for multiple admixture events involving Himalayan populations and South/East Asians between 200 and 2,000 years ago. In comparisons with available ancient genomes, the Himalayans, like other East and South Asian populations, showed similar genetic affinity to Eurasian hunter-gatherers (a 24,000-year-old Upper Palaeolithic Siberian), and the related Bronze Age Yamnaya. The high-altitude Himalayan populations all shared a specific ancestral component, suggesting that genetic adaptation to life at high altitude originated only once in this region and subsequently spread. Combining four approaches to identifying specific positively selected loci, we confirmed that the strongest signals of high-altitude adaptation were located near the Endothelial PAS domain-containing protein 1 and Egl-9 Family Hypoxia Inducible Factor 1 loci, and discovered eight additional robust signals of high-altitude adaptation, five of which have strong biological functional links to such adaptation. In conclusion, the demographic history of Himalayan populations is complex, with strong local differentiation, reflecting both genetic and cultural factors; these populations also display evidence of multiple genetic adaptations to high-altitude environments.
Figures

References
-
- Aldenderfer M. 2011. Peopling the Tibetan plateau: insights from archaeology. High Alt Med Biol. 122:141–147. - PubMed
-
- Allentoft ME, Sikora M, Sjogren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlstrom T, Vinner L, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 5227555:167–172. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

