High parasite diversity accelerates host adaptation and diversification
- PMID: 29798882
- PMCID: PMC7612933
- DOI: 10.1126/science.aam9974
High parasite diversity accelerates host adaptation and diversification
Abstract
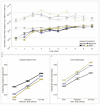
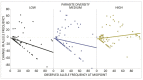
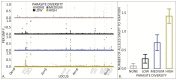
Host-parasite species pairs are known to coevolve, but how multiple parasites coevolve with their host is unclear. By using experimental coevolution of a host bacterium and its viral parasites, we revealed that diverse parasite communities accelerated host evolution and altered coevolutionary dynamics to enhance host resistance and decrease parasite infectivity. Increases in parasite diversity drove shifts in the mode of selection from fluctuating (Red Queen) dynamics to predominately directional (arms race) dynamics. Arms race dynamics were characterized by selective sweeps of generalist resistance mutations in the genes for the host bacterium's cell surface lipopolysaccharide (a bacteriophage receptor), which caused faster molecular evolution within host populations and greater genetic divergence among populations. These results indicate that exposure to multiple parasites influences the rate and type of host-parasite coevolution.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

References
-
- Haldane JBS. Disease and evolution. La Ric Sci. 1949;19:68–76.
-
- Bell G. The masterpiece of nature : the evolution and genetics of sexuality. Croom Helm; London: 1982.
-
- Hamilton WD. Sex versus Non-Sex versus Parasite. Oikos. 1980;35:282.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

