Characterization of Hemagglutinin Antigens on Influenza Virus and within Vaccines Using Electron Microscopy
- PMID: 29799445
- PMCID: PMC6027289
- DOI: 10.3390/vaccines6020031
Characterization of Hemagglutinin Antigens on Influenza Virus and within Vaccines Using Electron Microscopy
Abstract
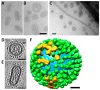
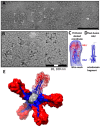
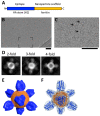
Influenza viruses affect millions of people worldwide on an annual basis. Although vaccines are available, influenza still causes significant human mortality and morbidity. Vaccines target the major influenza surface glycoprotein hemagglutinin (HA). However, circulating HA subtypes undergo continual variation in their dominant epitopes, requiring vaccines to be updated annually. A goal of next-generation influenza vaccine research is to produce broader protective immunity against the different types, subtypes, and strains of influenza viruses. One emerging strategy is to focus the immune response away from variable epitopes, and instead target the conserved stem region of HA. To increase the display and immunogenicity of the HA stem, nanoparticles are being developed to display epitopes in a controlled spatial arrangement to improve immunogenicity and elicit protective immune responses. Engineering of these nanoparticles requires structure-guided design to optimize the fidelity and valency of antigen presentation. Here, we review electron microscopy applied to study the 3D structures of influenza viruses and different vaccine antigens. Structure-guided information from electron microscopy should be integrated into pipelines for the development of both more efficacious seasonal and universal influenza vaccine antigens. The lessons learned from influenza vaccine electron microscopic research could aid in the development of novel vaccines for other pathogens.
Keywords: cryo-EM; design; electron microscopy; influenza; structure; vaccines.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

