PaintOmics 3: a web resource for the pathway analysis and visualization of multi-omics data
- PMID: 29800320
- PMCID: PMC6030972
- DOI: 10.1093/nar/gky466
PaintOmics 3: a web resource for the pathway analysis and visualization of multi-omics data
Abstract
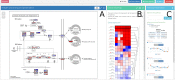
The increasing availability of multi-omic platforms poses new challenges to data analysis. Joint visualization of multi-omics data is instrumental in better understanding interconnections across molecular layers and in fully utilizing the multi-omic resources available to make biological discoveries. We present here PaintOmics 3, a web-based resource for the integrated visualization of multiple omic data types onto KEGG pathway diagrams. PaintOmics 3 combines server-end capabilities for data analysis with the potential of modern web resources for data visualization, providing researchers with a powerful framework for interactive exploration of their multi-omics information. Unlike other visualization tools, PaintOmics 3 covers a comprehensive pathway analysis workflow, including automatic feature name/identifier conversion, multi-layered feature matching, pathway enrichment, network analysis, interactive heatmaps, trend charts, and more. It accepts a wide variety of omic types, including transcriptomics, proteomics and metabolomics, as well as region-based approaches such as ATAC-seq or ChIP-seq data. The tool is freely available at www.paintomics.org.
Figures




References
-
- Cavill R., Jennen D., Kleinjans J., Briedé J.J.. Transcriptomic and metabolomic data integration. Brief. Bioinform. 2015; 17:891–901. - PubMed
-
- Bastian M., Heymann S., Jacomy M. et al. . Gephi: an open source software for exploring and manipulating networks. ICWSM. 2009; 8:361–362.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

