Agmatine Production by Aspergillus oryzae Is Elevated by Low pH during Solid-State Cultivation
- PMID: 29802188
- PMCID: PMC6052267
- DOI: 10.1128/AEM.00722-18
Agmatine Production by Aspergillus oryzae Is Elevated by Low pH during Solid-State Cultivation
Abstract
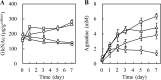
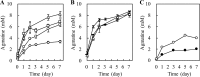
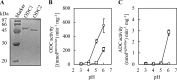
Sake (rice wine) produced by multiple parallel fermentation (MPF) involving Aspergillus oryzae (strain RW) and Saccharomyces cerevisiae under solid-state cultivation conditions contained 3.5 mM agmatine, while that produced from enzymatically saccharified rice syrup by S. cerevisiae contained <0.01 mM agmatine. Agmatine was also produced in ethanol-free rice syrup prepared with A. oryzae under solid-state cultivation (3.1 mM) but not under submerged cultivation, demonstrating that A. oryzae in solid-state culture produces agmatine. The effect of cultivation conditions on agmatine production was examined. Agmatine production was boosted at 30°C and reached the highest level (6.3 mM) at pH 5.3. The addition of l-lactic, succinic, and citric acids reduced the initial culture pHs to 3.0, 3.5, and 3.2, respectively, resulting in a further increase in agmatine accumulation (8.2, 8.7, and 8.3 mM, respectively). Homogenate from a solid-state culture exhibited a maximum l-arginine decarboxylase (ADC) activity (74 pmol · min-1 · μg-1) at pH 3.0 at 30°C; homogenate from a submerged culture exhibited an extremely low activity (<0.3 pmol · min-1 · μg-1) under all conditions tested. These observations indicated that efficient agmatine production in ethanol-free rice syrup is achieved by an unidentified low-pH-dependent ADC induced during solid-state cultivation of A. oryzae, even though A. oryzae lacks ADC orthologs and instead possesses four ornithine decarboxylases (ODC1 to ODC4). Recombinant ODC1 and ODC2 exhibited no ADC activity at acidic pH (pH < 4.0), suggesting that other decarboxylases or an unidentified ADC is involved in agmatine production.IMPORTANCE It has been speculated that, in general, fungi do not synthesize agmatine from l-arginine because they do not possess genes encoding arginine decarboxylase. Numerous preclinical studies have shown that agmatine exerts pleiotropic effects on various molecular targets, leading to an improved quality of life. In the present study, we first demonstrated that l-arginine was a feasible substrate for agmatine production by the fungus Aspergillus oryzae RW. We observed that the productivity of agmatine by A. oryzae RW was elevated at low pH only during solid-state cultivation. A. oryzae is utilized in the production of various Asian fermented foods. The saccharification conditions optimized in the current study could be employed not only in the production of an agmatine-containing ethanol-free rice syrup but also in the production of many types of fermented foods, such as soy sauce (shoyu), rice vinegar, etc., as well as for use as novel therapeutic agents and nutraceuticals.
Keywords: Aspergillus oryzae; agmatine; polyamine; rice syrup; saccharification.
Copyright © 2018 American Society for Microbiology.
Figures




References
-
- Kusano T, Suzuki H. 2015. Polyamines: a universal molecular nexus for growth, survival, and specialized metabolism. Springer Japan, Tokyo, Japan.
-
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Fröhlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. 2009. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11:1305–1314. doi:10.1038/ncb1975. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

