The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell
- PMID: 29802201
- PMCID: PMC6052207
- DOI: 10.1074/jbc.RA118.002715
The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell
Abstract
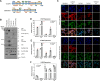
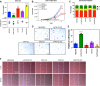
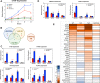
The Hippo pathway plays an important role in regulating tissue homeostasis, and its effectors, the transcriptional co-activators Yes-associated protein (YAP) and WW domain-containing transcription regulator 1 (WWTR1 or TAZ), are responsible for mediating the vast majority of its physiological functions. Although YAP and TAZ are thought to be largely redundant and similarly regulated by Hippo signaling, they have developmental, structural, and physiological differences that suggest they may differ in their regulation and downstream functions. To better understand the functions of YAP and TAZ in the Hippo pathway, using CRISPR/Cas9, we generated YAP KO, TAZ KO, and YAP/TAZ KO cell lines in HEK293A cells. We evaluated them in response to many environmental conditions and stimuli and used RNA-Seq to compare their transcriptional profiles. We found that YAP inactivation has a greater effect on cellular physiology (namely, cell spreading, volume, granularity, glucose uptake, proliferation, and migration) than TAZ inactivation. However, functional redundancy between YAP and TAZ was also observed. In summary, our findings confirm that the Hippo pathway effectors YAP and TAZ are master regulators for multiple cellular processes but also reveal that YAP has a stronger influence than TAZ.
Keywords: CRISPR/Cas; Hippo pathway; TAZ; Yes-associated protein (YAP); cell biology; gene knockout.
© 2018 Plouffe et al.
Conflict of interest statement
K.-L. G. is a co-founder of and has an equity interest in Vivace Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures



References
-
- Hong J. H., Hwang E. S., McManus M. T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B. M., Sharp P. A., Hopkins N., and Yaffe M. B. (2005) TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309, 1074–1078 10.1126/science.1110955 - DOI - PubMed
-
- Kanai F., Marignani P. A., Sarbassova D., Yagi R., Hall R. A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L. C., and Yaffe M. B. (2000) TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19, 6778–6791 10.1093/emboj/19.24.6778 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

