The Chromatin Remodelers PKL and PIE1 Act in an Epigenetic Pathway That Determines H3K27me3 Homeostasis in Arabidopsis
- PMID: 29802212
- PMCID: PMC6048792
- DOI: 10.1105/tpc.17.00867
The Chromatin Remodelers PKL and PIE1 Act in an Epigenetic Pathway That Determines H3K27me3 Homeostasis in Arabidopsis
Abstract
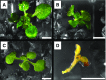
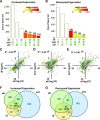
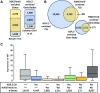
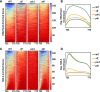
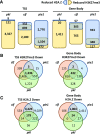
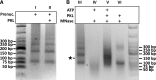
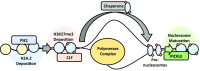
Selective, tissue-specific gene expression is facilitated by the epigenetic modification H3K27me3 (trimethylation of lysine 27 on histone H3) in plants and animals. Much remains to be learned about how H3K27me3-enriched chromatin states are constructed and maintained. Here, we identify a genetic interaction in Arabidopsis thaliana between the chromodomain helicase DNA binding chromatin remodeler PICKLE (PKL), which promotes H3K27me3 enrichment, and the SWR1-family remodeler PHOTOPERIOD INDEPENDENT EARLY FLOWERING1 (PIE1), which incorporates the histone variant H2A.Z. Chromatin immunoprecipitation-sequencing and RNA-sequencing reveal that PKL, PIE1, and the H3K27 methyltransferase CURLY LEAF act in a common gene expression pathway and are required for H3K27me3 levels genome-wide. Additionally, H3K27me3-enriched genes are largely a subset of H2A.Z-enriched genes, further supporting the functional linkage between these marks. We also found that recombinant PKL acts as a prenucleosome maturation factor, indicating that it promotes retention of H3K27me3. These data support the existence of an epigenetic pathway in which PIE1 promotes H2A.Z, which in turn promotes H3K27me3 deposition. After deposition, PKL promotes retention of H3K27me3 after DNA replication and/or transcription. Our analyses thus reveal roles for H2A.Z and ATP-dependent remodelers in construction and maintenance of H3K27me3-enriched chromatin in plants.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures



Similar articles
-
The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27.Plant Physiol. 2012 May;159(1):418-32. doi: 10.1104/pp.112.194878. Epub 2012 Mar 27. Plant Physiol. 2012. PMID: 22452853 Free PMC article.
-
SWR1 Chromatin-Remodeling Complex Subunits and H2A.Z Have Non-overlapping Functions in Immunity and Gene Regulation in Arabidopsis.Mol Plant. 2016 Jul 6;9(7):1051-65. doi: 10.1016/j.molp.2016.04.003. Epub 2016 Apr 27. Mol Plant. 2016. PMID: 27131447 Free PMC article.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
-
Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z.Plant Cell. 2007 Jan;19(1):74-83. doi: 10.1105/tpc.106.048447. Epub 2007 Jan 12. Plant Cell. 2007. PMID: 17220196 Free PMC article.
-
The beauty of being a variant: H2A.Z and the SWR1 complex in plants.Mol Plant. 2009 Jul;2(4):565-577. doi: 10.1093/mp/ssp019. Epub 2009 Mar 31. Mol Plant. 2009. PMID: 19825639 Review.
Cited by
-
Low-Phosphate Chromatin Dynamics Predict a Cell Wall Remodeling Network in Rice Shoots.Plant Physiol. 2020 Mar;182(3):1494-1509. doi: 10.1104/pp.19.01153. Epub 2019 Dec 19. Plant Physiol. 2020. PMID: 31857425 Free PMC article.
-
Advances in biological functions and mechanisms of histone variants in plants.Front Genet. 2023 Jul 31;14:1229782. doi: 10.3389/fgene.2023.1229782. eCollection 2023. Front Genet. 2023. PMID: 37588047 Free PMC article. Review.
-
A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding.EMBO J. 2020 Apr 1;39(7):e102008. doi: 10.15252/embj.2019102008. Epub 2020 Mar 2. EMBO J. 2020. PMID: 32115743 Free PMC article.
-
PICKLE RELATED 2 is a Neofunctionalized Gene Duplicate Under Positive Selection With Antagonistic Effects to the Ancestral PICKLE Gene on the Seed Transcriptome.Genome Biol Evol. 2023 Nov 1;15(11):evad191. doi: 10.1093/gbe/evad191. Genome Biol Evol. 2023. PMID: 37931037 Free PMC article.
-
The histone variant H2A.W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation.Nat Commun. 2021 May 11;12(1):2683. doi: 10.1038/s41467-021-22993-5. Nat Commun. 2021. PMID: 33976212 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

