Response to Persistent ER Stress in Plants: A Multiphasic Process That Transitions Cells from Prosurvival Activities to Cell Death
- PMID: 29802214
- PMCID: PMC6048783
- DOI: 10.1105/tpc.18.00153
Response to Persistent ER Stress in Plants: A Multiphasic Process That Transitions Cells from Prosurvival Activities to Cell Death
Abstract
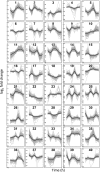
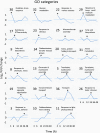
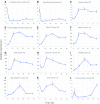
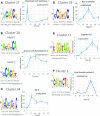
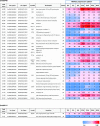
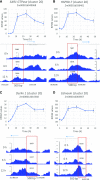
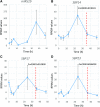
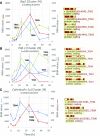
The unfolded protein response (UPR) is a highly conserved response that protects plants from adverse environmental conditions. The UPR is elicited by endoplasmic reticulum (ER) stress, in which unfolded and misfolded proteins accumulate within the ER. Here, we induced the UPR in maize (Zea mays) seedlings to characterize the molecular events that occur over time during persistent ER stress. We found that a multiphasic program of gene expression was interwoven among other cellular events, including the induction of autophagy. One of the earliest phases involved the degradation by regulated IRE1-dependent RNA degradation (RIDD) of RNA transcripts derived from a family of peroxidase genes. RIDD resulted from the activation of the promiscuous ribonuclease activity of ZmIRE1 that attacks the mRNAs of secreted proteins. This was followed by an upsurge in expression of the canonical UPR genes indirectly driven by ZmIRE1 due to its splicing of Zmbzip60 mRNA to make an active transcription factor that directly upregulates many of the UPR genes. At the peak of UPR gene expression, a global wave of RNA processing led to the production of many aberrant UPR gene transcripts, likely tempering the ER stress response. During later stages of ER stress, ZmIRE1's activity declined, as did the expression of survival modulating genes, Bax inhibitor1 and Bcl-2-associated athanogene7, amid a rising tide of cell death. Thus, in response to persistent ER stress, maize seedlings embark on a course of gene expression and cellular events progressing from adaptive responses to cell death.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

