Modeling the Embrace of a Mutator: APOBEC Selection of Nucleic Acid Ligands
- PMID: 29803538
- PMCID: PMC6073885
- DOI: 10.1016/j.tibs.2018.04.013
Modeling the Embrace of a Mutator: APOBEC Selection of Nucleic Acid Ligands
Abstract
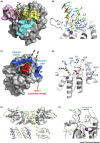
The 11-member APOBEC (apolipoprotein B mRNA editing catalytic polypeptide-like) family of zinc-dependent cytidine deaminases bind to RNA and single-stranded DNA (ssDNA) and, in specific contexts, modify select (deoxy)cytidines to (deoxy)uridines. In this review, we describe advances made through high-resolution co-crystal structures of APOBECs bound to mono- or oligonucleotides that reveal potential substrate-specific binding sites at the active site and non-sequence-specific nucleic acid binding sites distal to the active site. We also discuss the effect of APOBEC oligomerization on functionality. Future structural studies will need to address how ssDNA binding away from the active site may enhance catalysis and the mechanism by which RNA binding may modulate catalytic activity on ssDNA.
Keywords: APOBEC; RNA and DNA binding; RNA editing; cancer; crystal structure; cytidine deaminase; gene editing; innate and adaptive immunity; mutation; structural biology; zinc-dependent deaminase..
Copyright © 2018 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

