Mis16 Switches Function from a Histone H4 Chaperone to a CENP-ACnp1-Specific Assembly Factor through Eic1 Interaction
- PMID: 29804820
- PMCID: PMC6031460
- DOI: 10.1016/j.str.2018.04.012
Mis16 Switches Function from a Histone H4 Chaperone to a CENP-ACnp1-Specific Assembly Factor through Eic1 Interaction
Abstract
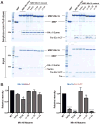
The Mis18 complex, composed of Mis16, Eic1, and Mis18 in fission yeast, selectively deposits the centromere-specific histone H3 variant, CENP-ACnp1, at centromeres. How the intact Mis18 holo-complex oligomerizes and how Mis16, a well-known ubiquitous histone H4 chaperone, plays a centromere-specific role in the Mis18 holo-complex, remain unclear. Here, we report the stoichiometry of the intact Mis18 holo-complex as (Mis16)2:(Eic1)2:(Mis18)4 using analytical ultracentrifugation. We further determine the crystal structure of Schizosaccharomyces pombe Mis16 in complex with the C-terminal portion of Eic1 (Eic1-CT). Notably, Mis16 accommodates Eic1-CT through the binding pocket normally occupied by histone H4, indicating that Eic1 and H4 compete for the same binding site, providing a mechanism for Mis16 to switch its binding partner from histone H4 to Eic1. Thus, our analyses not only determine the stoichiometry of the intact Mis18 holo-complex but also uncover the molecular mechanism by which Mis16 plays a centromere-specific role through Eic1 association.
Keywords: CENP-A; Eic1; Mis16; X-ray crystallography; analytical ultracentrifugation; centromere; fission yeast; histone chaperone; kinetochore; the Mis18 holo-complex.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

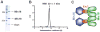




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

