CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning
- PMID: 29804837
- PMCID: PMC6026039
- DOI: 10.1016/j.cell.2018.04.035
CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning
Abstract
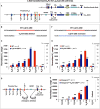
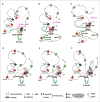
RAG endonuclease initiates antibody heavy chain variable region exon assembly from V, D, and J segments within a chromosomal V(D)J recombination center (RC) by cleaving between paired gene segments and flanking recombination signal sequences (RSSs). The IGCR1 control region promotes DJH intermediate formation by isolating Ds, JHs, and RCs from upstream VHs in a chromatin loop anchored by CTCF-binding elements (CBEs). How VHs access the DJHRC for VH to DJH rearrangement was unknown. We report that CBEs immediately downstream of frequently rearranged VH-RSSs increase recombination potential of their associated VH far beyond that provided by RSSs alone. This CBE activity becomes particularly striking upon IGCR1 inactivation, which allows RAG, likely via loop extrusion, to linearly scan chromatin far upstream. VH-associated CBEs stabilize interactions of D-proximal VHs first encountered by the DJHRC during linear RAG scanning and thereby promote dominant rearrangement of these VHs by an unanticipated chromatin accessibility-enhancing CBE function.
Keywords: 3C-HTGTS; CBE orientation; CTCF-binding elements; HTGTS-Rep-seq; RAG chromatin scanning; V(D)J recombination; V(H) accessibility; V(H)81X; intergenic control region 1; loop extrusion.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

