PDn-3 DPA Pathway Regulates Human Monocyte Differentiation and Macrophage Function
- PMID: 29805036
- PMCID: PMC6024030
- DOI: 10.1016/j.chembiol.2018.04.017
PDn-3 DPA Pathway Regulates Human Monocyte Differentiation and Macrophage Function
Abstract
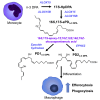
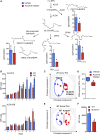
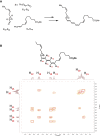
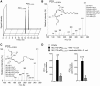
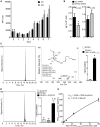
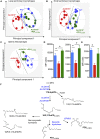
Macrophages are central in orchestrating the clearance of apoptotic cells and cellular debris during inflammation, with the mechanism(s) regulating this process remaining of interest. Herein, we found that the n-3 docosapentaenoic acid-derived protectin (PDn-3 DPA) biosynthetic pathway regulated the differentiation of human monocytes, altering macrophage phenotype, efferocytosis, and bacterial phagocytosis. Using lipid mediator profiling, human primary cells and recombinant enzymes we found that human 15-lipoxygenases initiate the PDn-3 DPA pathway catalyzing the formation of an allylic epoxide. The complete stereochemistry of this epoxide was determined using stereocontrolled total organic synthesis as 16S,17S-epoxy-7Z,10Z,12E,14E,19Z-docosapentaenoic acid (16S,17S-ePDn-3 DPA). This intermediate was enzymatically converted by epoxide hydrolases to PD1n-3 DPA and PD2n-3 DPA, with epoxide hydrolase 2 converting 16S,17S-ePDn-3 DPA to PD2n-3 DPA in human monocytes. Taken together these results establish the PDn-3 DPA biosynthetic pathway in human monocytes and macrophages and its role in regulating macrophage resolution responses.
Keywords: lipid mediators; omega-3; resolution; total organic synthesis.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Borgeat P., Fruteau de Laclos B., Picard S., Drapeau J., Vallerand P., Corey E.J. Studies on the mechanism of formation of the 5S, 12S-dihydroxy-6,8,10,14(E, Z,E,Z)-icosatetraenoic acid in leukocytes. Prostaglandins. 1982;23:713–724. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

