Key genes and pathways in measles and their interaction with environmental chemicals
- PMID: 29805511
- PMCID: PMC5958750
- DOI: 10.3892/etm.2018.6050
Key genes and pathways in measles and their interaction with environmental chemicals
Abstract
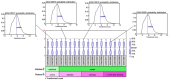
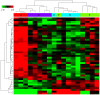
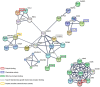
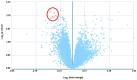
The aim of the present study was to explore key genes that may have a role in the pathology of measles virus infection and to clarify the interaction networks between environmental factors and differentially expressed genes (DEGs). After screening the database of the Gene Expression Omnibus of the National Center for Biotechnology Information, the dataset GSE5808 was downloaded and analyzed. A global normalization method was performed to minimize data inconsistencies and heterogeneity. DEGs during different stages of measles virus infection were explored using R software (v3.4.0). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the DEGs were performed using Cytoscape 3.4.0 software. A protein-protein interaction (PPI) network of the DEGs was obtained from the STRING database v9.05. A total of 43 DEGs were obtained from four analyzed sample groups, including 10 highly expressed genes and 33 genes with decreased expression. The most enriched pathways based on KEGG analysis were fatty acid elongation, cytokine-cytokine receptor interaction and RNA degradation. The genes mentioned in the PPI network were mainly associated with protein binding and chemokine activity. A total of 219 chemicals were identified that may, jointly or on their own, interact with the 6 DEGs between the control group and patients with measles (at hospital entry), including benzo(a)pyrene (BaP) and tetrachlorodibenzodioxin (TCDD). In conclusion, the present study revealed that chemokines and environmental chemicals, e.g. BaP and TCDD, may affect the development of measles.
Keywords: and measles; differentially expressed genes; environmental risk factors.
Figures

Similar articles
-
The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data.Med Oncol. 2017 Jun;34(6):101. doi: 10.1007/s12032-017-0963-9. Epub 2017 Apr 21. Med Oncol. 2017. PMID: 28432618 Free PMC article.
-
Analysis of expression profile data identifies key genes and pathways in hepatocellular carcinoma.Oncol Lett. 2018 Feb;15(2):2625-2630. doi: 10.3892/ol.2017.7534. Epub 2017 Dec 6. Oncol Lett. 2018. PMID: 29434983 Free PMC article.
-
Comprehensive Bioinformatics Analysis of the Immune Mechanism of Dendritic Cells Against Measles Virus.Med Sci Monit. 2019 Feb 1;25:903-912. doi: 10.12659/MSM.912949. Med Sci Monit. 2019. PMID: 30705250 Free PMC article.
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
Analysis of the protein-protein interaction networks of differentially expressed genes in pulmonary embolism.Mol Med Rep. 2015 Apr;11(4):2527-33. doi: 10.3892/mmr.2014.3006. Epub 2014 Nov 26. Mol Med Rep. 2015. PMID: 25434468 Free PMC article.
References
-
- World Health Organization, corp-author. http://www.who.int/mediacentre/factsheets/fs286/en/ [Sep 29;2017 ];Measles Fact Sheets. 2016
-
- Centers for Disease Control and Prevention (CDC): Measles-United States, 2011. MMWR Morb Mortal Wkly. Rep. 2012;61:253–257. - PubMed
-
- http://iris.wpro.who.int/bitstream/handle/10665.1/13539/Measles-Rubella-.... [Sep 29;2017 ];World Health Organization: Measles-Rubella Bulletin. 2017
-
- No authors listed: Measles vaccines: WHO position paper. Wkly Epidemiol Rec. 2009;84:349–360. (In English, French) - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
