Single-molecule analysis reveals the mechanism of transcription activation in M. tuberculosis
- PMID: 29806016
- PMCID: PMC5966222
- DOI: 10.1126/sciadv.aao5498
Single-molecule analysis reveals the mechanism of transcription activation in M. tuberculosis
Abstract
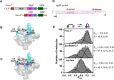
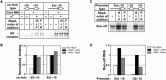
The σ subunit of bacterial RNA polymerase (RNAP) controls recognition of the -10 and -35 promoter elements during transcription initiation. Free σ adopts a "closed," or inactive, conformation incompatible with promoter binding. The conventional two-state model of σ activation proposes that binding to core RNAP induces formation of an "open," active, σ conformation, which is optimal for promoter recognition. Using single-molecule Förster resonance energy transfer, we demonstrate that vegetative-type σ subunits exist in open and closed states even after binding to the RNAP core. As an extreme case, RNAP from Mycobacterium tuberculosis preferentially retains σ in the closed conformation, which is converted to the open conformation only upon binding by the activator protein RbpA and interaction with promoter DNA. These findings reveal that the conformational dynamics of the σ subunit in the RNAP holoenzyme is a target for regulation by transcription factors and plays a critical role in promoter recognition.
Figures



References
-
- Feklístov A., Sharon B. D., Darst S. A., Gross C. A., Bacterial sigma factors: A historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376 (2014). - PubMed
-
- Dombroski A. J., Walter W. A., Gross C. A., Amino-terminal amino acids modulate σ-factor DNA-binding activity. Genes Dev. 7, 2446–2455 (1993). - PubMed
-
- Dombroski A. J., Walter W. A., Record M. T. Jr, Siegele D. A., Gross C. A., Polypeptides containing highly conserved regions of transcription initiation factor σ70 exhibit specificity of binding to promoter DNA. Cell 70, 501–512 (1992). - PubMed
-
- Callaci S., Heyduk E., Heyduk T., Core RNA polymerase from E. coli induces a major change in the domain arrangement of the σ70 subunit. Mol. Cell 3, 229–238 (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

