The interplay between microbes and the immune response in inflammatory bowel disease
- PMID: 29806140
- PMCID: PMC6117586
- DOI: 10.1113/JP275396
The interplay between microbes and the immune response in inflammatory bowel disease
Abstract
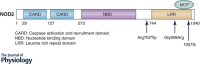
The aetiology and pathogenesis of inflammatory bowel disease (IBD) remains unclear but involves a complex interplay between genetic risk, environmental exposures, the immune system and the gut microbiota. Nearly two decades ago, the first susceptibility gene for Crohn's disease, NOD2, was identified within the IBD 1 locus. Since then, over 230 genetic risk loci have been associated with IBD and yet NOD2 remains the strongest association to date. As an intracellular innate immune sensor of bacteria, investigations into host-microbe interactions, involving both innate and adaptive immune responses, have become of particular interest in understanding the pathogenesis of IBD. Advancements in sequencing technology have lead to the groundbreaking characterization of the gut microbiota and its role in health and disease. While an altered microbiome has been described for IBD, whether it is a cause or an effect of the intestinal inflammation has yet to be determined. Moreover, the bidirectional relationship between the gut microbiota and the mucosal immune system adds to the multifaceted complexity of intestinal homeostasis. A better understanding of how host genetics, including NOD2, influence immune-microbe interactions and alter susceptibility to IBD is necessary in order to develop therapeutic and preventative treatments.
Keywords: NLRs; Nod2; adaptive immunity; gut microbiota; immune-microbe interactions; inflammatory bowel diseases; innate immunity.
© 2018 The Authors. The Journal of Physiology © 2018 The Physiological Society.
Figures
References
-
- Aranda R, Sydora BC, McAllister PL, Binder SW, Yang HY, Targan SR & Kronenberg M (1997). Analysis of intestinal lymphocytes in mouse colitis mediated by transfer of CD4+, CD45RBhigh T cells to SCID recipients. J Immunol 158, 3464–3473. - PubMed
-
- Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist‐Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, CHILD Study Investigators , Mohn WW, Turvey SE & Finlay BB (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7, 307ra152. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

