Murine HPV16 E7-expressing transgenic skin effectively emulates the cellular and molecular features of human high-grade squamous intraepithelial lesions
- PMID: 29807614
- PMCID: PMC5886957
- DOI: 10.1016/j.pvr.2017.10.001
Murine HPV16 E7-expressing transgenic skin effectively emulates the cellular and molecular features of human high-grade squamous intraepithelial lesions
Abstract
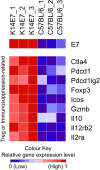
Currently available vaccines prevent HPV infection and development of HPV-associated malignancies, but do not cure existing HPV infections and dysplastic lesions. Persistence of infection(s) in immunocompetent patients may reflect induction of local immunosuppressive mechanisms by HPV, providing a target for therapeutic intervention. We have proposed that a mouse, expressing HPV16 E7 oncoprotein under a Keratin 14 promoter (K14E7 mice), and which develops epithelial hyperplasia, may assist with understanding local immune suppression mechanisms that support persistence of HPV oncogene-induced epithelial hyperplasia. K14E7 skin grafts recruit immune cells from immunocompetent hosts, but consistently fail to be rejected. Here, we review the literature on HPV-associated local immunoregulation, and compare the findings with published observations on the K14E7 transgenic murine model, including comparison of the transcriptome of human HPV-infected pre-malignancies with that of murine K14E7 transgenic skin. We argue from the similarity of i) the literature findings and ii) the transcriptome profiles that murine K14E7 transgenic skin recapitulates the cellular and secreted protein profiles of high-grade HPV-associated lesions in human subjects. We propose that the K14E7 mouse may be an appropriate model to further study the immunoregulatory effects of HPV E7 expression, and can facilitate development and testing of therapeutic vaccines.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures







References
-
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA: Cancer J. Clin. 2015;65:87–108. - PubMed
-
- Frazer I.H., Leggatt G.R., Mattarollo S.R. Prevention and treatment of papillomavirus-related cancers through immunization. Annu. Rev. Immunol. 2011;29:111–138. - PubMed
-
- Frazer I. Correlating immunity with protection for HPV infection. Int. J. Infect. Dis.: IJID: Off. Publ. Int. Soc. Infect. Dis. 2007;11(Suppl 2):S10–S16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

