Estrogen receptor subcellular localization and cardiometabolism
- PMID: 29807870
- PMCID: PMC6066739
- DOI: 10.1016/j.molmet.2018.05.009
Estrogen receptor subcellular localization and cardiometabolism
Abstract
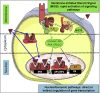
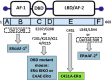
Background: In addition to their crucial role in reproduction, estrogens are key regulators of energy and glucose homeostasis and they also exert several cardiovascular protective effects. These beneficial actions are mainly mediated by estrogen receptor alpha (ERα), which is widely expressed in metabolic and vascular tissues. As a member of the nuclear receptor superfamily, ERα was primarily considered as a transcription factor that controls gene expression through the activation of its two activation functions (ERαAF-1 and ERαAF-2). However, besides these nuclear actions, a pool of ERα is localized in the vicinity of the plasma membrane, where it mediates rapid signaling effects called membrane-initiated steroid signals (MISS) that have been well described in vitro, especially in endothelial cells.
Scope of the review: This review aims to summarize our current knowledge of the mechanisms of nuclear vs membrane ERα activation that contribute to the cardiometabolic protection conferred by estrogens. Indeed, new transgenic mouse models (affecting either DNA binding, activation functions or membrane localization), together with the use of novel pharmacological tools that electively activate membrane ERα effects recently allowed to begin to unravel the different modes of ERα signaling in vivo.
Conclusion: Altogether, available data demonstrate the prominent role of ERα nuclear effects, and, more specifically, of ERαAF-2, in the preventive effects of estrogens against obesity, diabetes, and atheroma. However, membrane ERα signaling selectively mediates some of the estrogen endothelial/vascular effects (NO release, reendothelialization) and could also contribute to the regulation of energy balance, insulin sensitivity, and glucose metabolism. Such a dissection of ERα biological functions related to its subcellular localization will help to understand the mechanism of action of "old" ER modulators and to design new ones with an optimized benefit/risk profile.
Keywords: Cardiovascular system; Energy balance; Estrogen receptors; Genomic effects; Glucose homeostasis; Membrane-initiated steroid signals.
Copyright © 2018. Published by Elsevier GmbH.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

