Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs
- PMID: 29807989
- PMCID: PMC6055992
- DOI: 10.1038/s41557-018-0052-5
Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs
Abstract
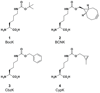
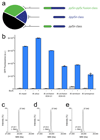
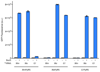
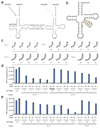
Genetically encoding distinct non-canonical amino acids (ncAAs) into proteins synthesized in cells requires mutually orthogonal aminoacyl-tRNA synthetase (aaRS)/tRNA pairs. The pyrrolysyl-tRNA synthetase/PyltRNA pair from Methanosarcina mazei (Mm) has been engineered to incorporate diverse ncAAs and is commonly considered an ideal pair for genetic code expansion. However, finding new aaRS/tRNA pairs that share the advantages of the MmPylRS/MmPyltRNA pair and are orthogonal to both endogenous aaRS/tRNA pairs and the MmPylRS/MmPyltRNA pair has proved challenging. Here we demonstrate that several ΔNPylRS/PyltRNACUA pairs, in which PylRS lacks an N-terminal domain, are active, orthogonal and efficiently incorporate ncAAs in Escherichia coli. We create new PylRS/PyltRNA pairs that are mutually orthogonal to the MmPylRS/MmPyltRNA pair and show that transplanting mutations that reprogram the ncAA specificity of MmPylRS into the new PylRS reprograms its substrate specificity. Finally, we show that distinct PylRS/PyltRNA-derived pairs can function in the same cell, decode distinct codons and incorporate distinct ncAAs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Optimizing orthogonality.Nat Chem. 2018 Aug;10(8):802-803. doi: 10.1038/s41557-018-0115-7. Nat Chem. 2018. PMID: 30030532 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

