RPA and RAD51: fork reversal, fork protection, and genome stability
- PMID: 29807999
- PMCID: PMC6006513
- DOI: 10.1038/s41594-018-0075-z
RPA and RAD51: fork reversal, fork protection, and genome stability
Abstract
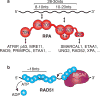
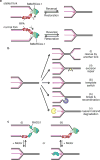
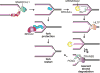
Replication protein A (RPA) and RAD51 are DNA-binding proteins that help maintain genome stability during DNA replication. These proteins regulate nucleases, helicases, DNA translocases, and signaling proteins to control replication, repair, recombination, and the DNA damage response. Their different DNA-binding mechanisms, enzymatic activities, and binding partners provide unique functionalities that cooperate to ensure that the appropriate activities are deployed at the right time to overcome replication challenges. Here we review and discuss the latest discoveries of the mechanisms by which these proteins work to preserve genome stability, with a focus on their actions in fork reversal and fork protection.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

