KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis
- PMID: 29808023
- PMCID: PMC7116356
- DOI: 10.1038/s41477-018-0160-7
KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis
Abstract
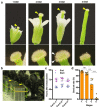
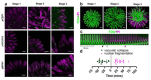
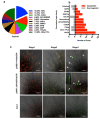
Flowers have a species-specific functional life span that determines the time window in which pollination, fertilization and seed set can occur. The stigma tissue plays a key role in flower receptivity by intercepting pollen and initiating pollen tube growth toward the ovary. In this article, we show that a developmentally controlled cell death programme terminates the functional life span of stigma cells in Arabidopsis. We identified the leaf senescence regulator ORESARA1 (also known as ANAC092) and the previously uncharacterized KIRA1 (also known as ANAC074) as partially redundant transcription factors that modulate stigma longevity by controlling the expression of programmed cell death-associated genes. KIRA1 expression is sufficient to induce cell death and terminate floral receptivity, whereas lack of both KIRA1 and ORESARA1 substantially increases stigma life span. Surprisingly, the extension of stigma longevity is accompanied by only a moderate extension of flower receptivity, suggesting that additional processes participate in the control of the flower's receptive life span.
Figures




Comment in
-
The stigma of death.Nat Plants. 2018 Jun;4(6):323-324. doi: 10.1038/s41477-018-0168-z. Nat Plants. 2018. PMID: 29808022 No abstract available.
References
-
- Jones ML. Ethylene signaling is required for pollination-accelerated corolla senescence in petunias. Plant Science. 2008;175:190–196.
-
- Williams RR. The Effect of Summer Nitrogen Applications on The Quality of Apple Blossom. Journal of Horticultural Science. 1965;40:31–41.
-
- Primack RB. Longevity of Individual Flowers. Annual Review of Ecology and Systematics. 1985;16:15–37.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

