A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA
- PMID: 29809147
- PMCID: PMC6031429
- DOI: 10.7554/eLife.34100
A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA
Abstract
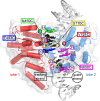
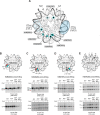
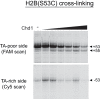
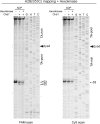
As superfamily 2 (SF2)-type translocases, chromatin remodelers are expected to use an inchworm-type mechanism to walk along DNA. Yet how they move DNA around the histone core has not been clear. Here we show that a remodeler ATPase motor can shift large segments of DNA by changing the twist and length of nucleosomal DNA at superhelix location 2 (SHL2). Using canonical and variant 601 nucleosomes, we find that the Saccharomyces cerevisiae Chd1 remodeler decreased DNA twist at SHL2 in nucleotide-free and ADP-bound states, and increased twist with transition state analogs. These differences in DNA twist allow the open state of the ATPase to pull in ~1 base pair (bp) by stabilizing a small DNA bulge, and closure of the ATPase to shift the DNA bulge toward the dyad. We propose that such formation and elimination of twist defects underlie the mechanism of nucleosome sliding by CHD-, ISWI-, and SWI/SNF-type remodelers.
Keywords: DNA translocation; S. cerevisiae; Snf2 ATPase; biochemistry; chemical biology; chromatin remodeling; chromosomes; gene expression; nucleosome; superfamily 2 ATPase; twist defect; xenopus.
© 2018, Winger et al.
Conflict of interest statement
JW, IN, RL, GB No competing interests declared
Figures













References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

