The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins
- PMID: 29809151
- PMCID: PMC5995541
- DOI: 10.7554/eLife.37018
The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins
Abstract
The endoplasmic reticulum (ER) supports biosynthesis of proteins with diverse transmembrane domain (TMD) lengths and hydrophobicity. Features in transmembrane domains such as charged residues in ion channels are often functionally important, but could pose a challenge during cotranslational membrane insertion and folding. Our systematic proteomic approaches in both yeast and human cells revealed that the ER membrane protein complex (EMC) binds to and promotes the biogenesis of a range of multipass transmembrane proteins, with a particular enrichment for transporters. Proximity-specific ribosome profiling demonstrates that the EMC engages clients cotranslationally and immediately following clusters of TMDs enriched for charged residues. The EMC can remain associated after completion of translation, which both protects clients from premature degradation and allows recruitment of substrate-specific and general chaperones. Thus, the EMC broadly enables the biogenesis of multipass transmembrane proteins containing destabilizing features, thereby mitigating the trade-off between function and stability.
Keywords: EMC; cell biology; endoplasmic reticulum; human; ion channel; transmembrane; transporter.
© 2018, Shurtleff et al.
Conflict of interest statement
MS, DI, JH, NS, EC, MJ, JW, KP, CJ, PS, SV, HH, JC, AB, JB, AF, GB, JW No competing interests declared
Figures
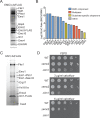
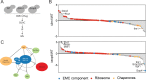






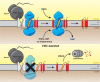
References
-
- Aviram N, Ast T, Costa EA, Arakel EC, Chuartzman SG, Jan CH, Haßdenteufel S, Dudek J, Jung M, Schorr S, Zimmermann R, Schwappach B, Weissman JS, Schuldiner M. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 2016;540:134–138. doi: 10.1038/nature20169. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103481/GM/NIGMS NIH HHS/United States
- AG041826/NH/NIH HHS/United States
- 1DP2GM110772-01/NH/NIH HHS/United States
- S10 OD016229/OD/NIH HHS/United States
- GM075061/NH/NIH HHS/United States
- Faculty Scholar Grant/HHMI/Howard Hughes Medical Institute/United States
- R01 AG041826/AG/NIA NIH HHS/United States
- R01 GM075061/GM/NIGMS NIH HHS/United States
- ERC2012-SyG_318987-ToPAG/European Research Council/International
- 8P41GM103481/NH/NIH HHS/United States
- 1S10OD16229/NH/NIH HHS/United States
- DP2 GM110772/GM/NIGMS NIH HHS/United States
- Investigator Program/HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

