SuperDCA for genome-wide epistasis analysis
- PMID: 29813016
- PMCID: PMC6096938
- DOI: 10.1099/mgen.0.000184
SuperDCA for genome-wide epistasis analysis
Abstract
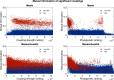
The potential for genome-wide modelling of epistasis has recently surfaced given the possibility of sequencing densely sampled populations and the emerging families of statistical interaction models. Direct coupling analysis (DCA) has previously been shown to yield valuable predictions for single protein structures, and has recently been extended to genome-wide analysis of bacteria, identifying novel interactions in the co-evolution between resistance, virulence and core genome elements. However, earlier computational DCA methods have not been scalable to enable model fitting simultaneously to 104-105 polymorphisms, representing the amount of core genomic variation observed in analyses of many bacterial species. Here, we introduce a novel inference method (SuperDCA) that employs a new scoring principle, efficient parallelization, optimization and filtering on phylogenetic information to achieve scalability for up to 105 polymorphisms. Using two large population samples of Streptococcus pneumoniae, we demonstrate the ability of SuperDCA to make additional significant biological findings about this major human pathogen. We also show that our method can uncover signals of selection that are not detectable by genome-wide association analysis, even though our analysis does not require phenotypic measurements. SuperDCA, thus, holds considerable potential in building understanding about numerous organisms at a systems biological level.
Keywords: epistasis; linkage disequilibrium; population genomics.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

