The small RNA complement of adult Schistosoma haematobium
- PMID: 29813122
- PMCID: PMC5993326
- DOI: 10.1371/journal.pntd.0006535
The small RNA complement of adult Schistosoma haematobium
Abstract
Background: Blood flukes of the genus Schistosoma cause schistosomiasis-a neglected tropical disease (NTD) that affects more than 200 million people worldwide. Studies of schistosome genomes have improved our understanding of the molecular biology of flatworms, but most of them have focused largely on protein-coding genes. Small non-coding RNAs (sncRNAs) have been explored in selected schistosome species and are suggested to play essential roles in the post-transcriptional regulation of genes, and in modulating flatworm-host interactions. However, genome-wide small RNA data are currently lacking for key schistosomes including Schistosoma haematobium-the causative agent of urogenital schistosomiasis of humans.
Methodology: MicroRNAs (miRNAs) and other sncRNAs of male and female adults of S. haematobium and small RNA transcription levels were explored by deep sequencing, genome mapping and detailed bioinformatic analyses.
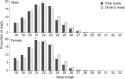
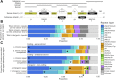
Principal findings: In total, 89 transcribed miRNAs were identified in S. haematobium-a similar complement to those reported for the congeners S. mansoni and S. japonicum. Of these miRNAs, 34 were novel, with no homologs in other schistosomes. Most miRNAs (n = 64) exhibited sex-biased transcription, suggestive of roles in sexual differentiation, pairing of adult worms and reproductive processes. Of the sncRNAs that were not miRNAs, some related to the spliceosome (n = 21), biogenesis of other RNAs (n = 3) or ribozyme functions (n = 16), whereas most others (n = 3798) were novel ('orphans') with unknown functions.
Conclusions: This study provides the first genome-wide sncRNA resource for S. haematobium, extending earlier studies of schistosomes. The present work should facilitate the future curation and experimental validation of sncRNA functions in schistosomes to enhance our understanding of post-transcriptional gene regulation and of the roles that sncRNAs play in schistosome reproduction, development and parasite-host cross-talk.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014;383(9936):2253–64. doi: 10.1016/S0140-6736(13)61949-2 . - DOI - PMC - PubMed
-
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6(7):411–25. Epub 2006/06/23. doi: 10.1016/S1473-3099(06)70521-7 . - DOI - PubMed
-
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460(7253):352–8. doi: 10.1038/nature08160 ; PubMed Central PMCID: PMC2756445. - DOI - PMC - PubMed
-
- Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460(7253):345–51. doi: 10.1038/nature08140 ; PubMed Central PMCID: PMC3747554. - DOI - PMC - PubMed
-
- Young ND, Jex AR, Li B, Liu S, Yang L, Xiong Z, et al. Whole-genome sequence of Schistosoma haematobium. Nat Genet. 2012;44(2):221–5. Epub 2012/01/17. doi: 10.1038/ng.1065 . - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

