Peptide-Programmable Nanoparticle Superstructures with Tailored Electrocatalytic Activity
- PMID: 29842775
- PMCID: PMC6556112
- DOI: 10.1021/acsnano.8b01146
Peptide-Programmable Nanoparticle Superstructures with Tailored Electrocatalytic Activity
Abstract
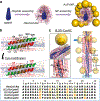
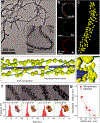
Biomaterials derived via programmable supramolecular protein assembly provide a viable means of constructing precisely defined structures. Here, we present programmed superstructures of AuPt nanoparticles (NPs) on carbon nanotubes (CNTs) that exhibit distinct electrocatalytic activities with respect to the nanoparticle positions via rationally modulated peptide-mediated assembly. De novo designed peptides assemble into six-helix bundles along the CNT axis to form a suprahelical structure. Surface cysteine residues of the peptides create AuPt-specific nucleation site, which allow for precise positioning of NPs onto helical geometries, as confirmed by 3-D reconstruction using electron tomography. The electrocatalytic model system, i.e., AuPt for oxygen reduction, yields electrochemical response signals that reflect the controlled arrangement of NPs in the intended assemblies. Our design approach can be expanded to versatile fields to build sophisticated functional assemblies.
Keywords: artificialy designed peptide; electrocatalytic oxygen reduction; electron tomography; nanoparticle superstructure; peptide-based catalyst; peptide-based superstructure 3-D reconstruction; supramolecular protein self-assembly.
Figures


References
-
- Busseron E; Ruff Y; Moulin E; Giuseppone N Supra-molecular Self-assemblies as Functional Nanomaterials. Nanoscale 2013, 5, 7098–7140. - PubMed
-
- Zhang SG Fabrication of Novel Biomaterials through Molecular Self-assembly. Nat. Biotechnol. 2003, 21, 1171–1178. - PubMed
-
- Chen CL; Rosi NL Preparation of Unique 1-D Nanoparticle Superstructures and Tailoring their Structural Features. J. Am. Chem. Soc. 2010, 132, 6902–6903. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

