An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray
- PMID: 29843789
- PMCID: PMC5975716
- DOI: 10.1186/s13059-018-1448-7
An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray
Abstract
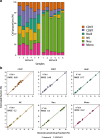
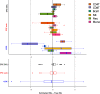
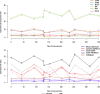
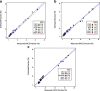
Genome-wide methylation arrays are powerful tools for assessing cell composition of complex mixtures. We compare three approaches to select reference libraries for deconvoluting neutrophil, monocyte, B-lymphocyte, natural killer, and CD4+ and CD8+ T-cell fractions based on blood-derived DNA methylation signatures assayed using the Illumina HumanMethylationEPIC array. The IDOL algorithm identifies a library of 450 CpGs, resulting in an average R2 = 99.2 across cell types when applied to EPIC methylation data collected on artificial mixtures constructed from the above cell types. Of the 450 CpGs, 69% are unique to EPIC. This library has the potential to reduce unintended technical differences across array platforms.
Keywords: Adults; B-cells; Cytotoxic T-lymphocytes; DNA methylation; Epigenetics; Helper T-cells; Leukocytes; Monocytes; Natural killer cells; Neutrophils.
Conflict of interest statement
Ethics approval and consent to participate
Cells used in these experiments were obtained commercially. All donors are anonymous. All the subjects provided written informed consent before donation to the commercial houses which provided the commercial cells.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, et al. Small-magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: the Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environ Health Perspect. 2017;125:511–526. doi: 10.1289/EHP595. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- P50CA097257/CA/NCI NIH HHS/United States
- R01CA207110/CA/NCI NIH HHS/United States
- R01CA52689/CA/NCI NIH HHS/United States
- P20GM108189/GM/NIGMS NIH HHS/United States
- P20GM104416/GM/NIGMS NIH HHS/United States
- P20 GM103418/GM/NIGMS NIH HHS/United States
- R01 CA216265/CA/NCI NIH HHS/United States
- R01 CA207110/CA/NCI NIH HHS/United States
- P50 CA097257/CA/NCI NIH HHS/United States
- R01CA216265/CA/NCI NIH HHS/United States
- R01 DE022772/DE/NIDCR NIH HHS/United States
- R01 CA207360/CA/NCI NIH HHS/United States
- R01DE022772/DE/NIDCR NIH HHS/United States
- P20GM103418/GM/NIGMS NIH HHS/United States
- R01 CA052689/CA/NCI NIH HHS/United States
- P20 GM104416/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

