Differential but Complementary HIF1α and HIF2α Transcriptional Regulation
- PMID: 29843956
- PMCID: PMC6036226
- DOI: 10.1016/j.ymthe.2018.05.004
Differential but Complementary HIF1α and HIF2α Transcriptional Regulation
Abstract
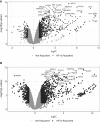
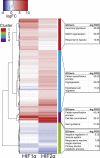
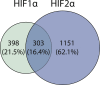
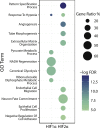
Effective vascular regeneration could provide therapeutic benefit for multiple pathologies, especially in chronic peripheral artery disease (PAD) and myocardial ischemia. The hypoxia inducible factors (HIFs) mediate the cellular transcriptional response to hypoxia and regulate multiple processes that are required for angiogenesis to ultimately restore perfusion and oxygen supply. In endothelial cells, both HIF1α and HIF2α are known to contribute to this role; however, the extent and individual roles of each of these HIFα remain unclear. To characterize the individual roles of HIFα, we sequenced the transcriptional outputs of stabilized forms of HIF1α and HIF2α, where they regulated 701 and 1,454 genes, respectively. HIF1α transcription primarily regulated metabolic reprogramming, whereas HIF2α exerted a larger role in regulating angiogenic extracellular signaling, guidance cues, and extracellular matrix remodeling factors. Furthermore, HIF2α almost exclusively regulated a large and diverse subset of transcription factors and coregulators that contribute to its diverse roles in hypoxia. Further understanding of how HIFs regulate cellular processes in hypoxia and angiogenesis could offer new avenues to modulate physiological angiogenesis to enhance revascularisation in ischemic conditions and other pathologies.
Keywords: EPAS1; HIF1a; HIF2a; RNA-seq; angiogenesis; cardiovascular disease; hypoxia; transcription; transcription factor.
Copyright © 2018 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Figures
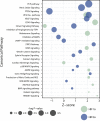


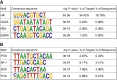
Similar articles
-
Enhanced HIF2α expression during human trophoblast differentiation into syncytiotrophoblast suppresses transcription of placental growth factor.Sci Rep. 2017 Sep 29;7(1):12455. doi: 10.1038/s41598-017-12685-w. Sci Rep. 2017. PMID: 28963486 Free PMC article.
-
A macrophage-dominant PI3K isoform controls hypoxia-induced HIF1α and HIF2α stability and tumor growth, angiogenesis, and metastasis.Mol Cancer Res. 2014 Oct;12(10):1520-31. doi: 10.1158/1541-7786.MCR-13-0682. Epub 2014 Aug 7. Mol Cancer Res. 2014. PMID: 25103499
-
Hypoxia inducible factors regulate the transcription of the sprouty2 gene and expression of the sprouty2 protein.PLoS One. 2017 Feb 14;12(2):e0171616. doi: 10.1371/journal.pone.0171616. eCollection 2017. PLoS One. 2017. PMID: 28196140 Free PMC article.
-
Drug resistance and cancer stem cells: the shared but distinct roles of hypoxia-inducible factors HIF1α and HIF2α.Clin Exp Pharmacol Physiol. 2017 Feb;44(2):153-161. doi: 10.1111/1440-1681.12693. Clin Exp Pharmacol Physiol. 2017. PMID: 27809360 Review.
-
Regulation of hypoxia-inducible gene expression after HIF activation.Exp Cell Res. 2017 Jul 15;356(2):182-186. doi: 10.1016/j.yexcr.2017.03.013. Epub 2017 Mar 9. Exp Cell Res. 2017. PMID: 28286304 Review.
Cited by
-
YAP/TAZ activation predicts clinical outcomes in mesothelioma and is conserved in in vitro model of driver mutations.Clin Transl Med. 2023 Feb;13(2):e1190. doi: 10.1002/ctm2.1190. Clin Transl Med. 2023. PMID: 36740402 Free PMC article.
-
HIF2α activation and mitochondrial deficit due to iron chelation cause retinal atrophy.EMBO Mol Med. 2023 Feb 8;15(2):e16525. doi: 10.15252/emmm.202216525. Epub 2023 Jan 16. EMBO Mol Med. 2023. PMID: 36645044 Free PMC article.
-
Targeting HIF-2α in the Tumor Microenvironment: Redefining the Role of HIF-2α for Solid Cancer Therapy.Cancers (Basel). 2022 Feb 28;14(5):1259. doi: 10.3390/cancers14051259. Cancers (Basel). 2022. PMID: 35267567 Free PMC article. Review.
-
Revisiting the HIF switch in the tumor and its immune microenvironment.Trends Cancer. 2022 Jan;8(1):28-42. doi: 10.1016/j.trecan.2021.10.004. Epub 2021 Nov 4. Trends Cancer. 2022. PMID: 34743924 Free PMC article. Review.
-
Disruption of adipocyte HIF-1α improves atherosclerosis through the inhibition of ceramide generation.Acta Pharm Sin B. 2022 Apr;12(4):1899-1912. doi: 10.1016/j.apsb.2021.10.001. Epub 2021 Oct 14. Acta Pharm Sin B. 2022. PMID: 35847503 Free PMC article.
References
-
- Bosch-Marce M., Okuyama H., Wesley J.B., Sarkar K., Kimura H., Liu Y.V., Zhang H., Strazza M., Rey S., Savino L. Effects of aging and hypoxia-inducible factor-1 activity on angiogenic cell mobilization and recovery of perfusion after limb ischemia. Circ. Res. 2007;101:1310–1318. - PubMed
-
- Kaelin W.G., Jr., Ratcliffe P.J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

