The Evolution of Molecular Compatibility between Bacteriophage ΦX174 and its Host
- PMID: 29844443
- PMCID: PMC5974221
- DOI: 10.1038/s41598-018-25914-7
The Evolution of Molecular Compatibility between Bacteriophage ΦX174 and its Host
Abstract
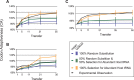
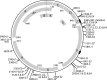
Viruses rely upon their hosts for biosynthesis of viral RNA, DNA and protein. This dependency frequently engenders strong selection for virus genome compatibility with potential hosts, appropriate gene regulation and expression necessary for a successful infection. While bioinformatic studies have shown strong correlations between codon usage in viral and host genomes, the selective factors by which this compatibility evolves remain a matter of conjecture. Engineered to include codons with a lesser usage and/or tRNA abundance within the host, three different attenuated strains of the bacterial virus ФX174 were created and propagated via serial transfers. Molecular sequence data indicate that biosynthetic compatibility was recovered rapidly. Extensive computational simulations were performed to assess the role of mutational biases as well as selection for translational efficiency in the engineered phage. Using bacteriophage as a model system, we can begin to unravel the evolutionary processes shaping codon compatibility between viruses and their host.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

