Asymmetric distribution of biomolecules of maternal origin in the Xenopus laevis egg and their impact on the developmental plan
- PMID: 29844480
- PMCID: PMC5974320
- DOI: 10.1038/s41598-018-26592-1
Asymmetric distribution of biomolecules of maternal origin in the Xenopus laevis egg and their impact on the developmental plan
Abstract
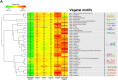
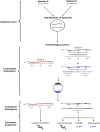
Asymmetric cell division is a ubiquitous feature during the development of higher organisms. Asymmetry is achieved by differential localization or activities of biological molecules such as proteins, and coding and non-coding RNAs. Here, we present subcellular transcriptomic and proteomic analyses along the animal-vegetal axis of Xenopus laevis eggs. More than 98% of the maternal mRNAs could be categorized into four localization profile groups: animal, vegetal, extremely vegetal, and a newly described group of mRNAs that we call extremely animal, which are mRNAs enriched in the animal cortex region. 3'UTRs of localized mRNAs were analyzed for localization motifs. Several putative motifs were discovered for vegetal and extremely vegetal mRNAs, while no distinct conserved motifs for the extremely animal mRNAs were identified, suggesting different localization mechanisms. Asymmetric profiles were also found for proteins, with correlation to those of corresponding mRNAs. Based on unexpected observation of the profiles of the homoeologous genes exd2 we propose a possible mechanism of genetic evolution.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

