ANXA11 mutations prevail in Chinese ALS patients with and without cognitive dementia
- PMID: 29845112
- PMCID: PMC5963931
- DOI: 10.1212/NXG.0000000000000237
ANXA11 mutations prevail in Chinese ALS patients with and without cognitive dementia
Abstract
Objective: To investigate the genetic contribution of ANXA11, a gene associated with amyotrophic lateral sclerosis (ALS), in Chinese ALS patients with and without cognitive dementia.
Methods: Sequencing all the coding exons of ANXA11 and intron-exon boundaries in 18 familial amyotrophic lateral sclerosis (FALS), 353 unrelated sporadic amyotrophic lateral sclerosis (SALS), and 12 Chinese patients with ALS-frontotemporal lobar dementia (ALS-FTD). The transcripts in peripheral blood generated from a splicing mutation were examined by reverse transcriptase PCR.
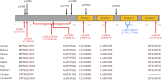
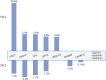
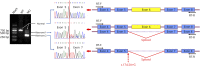
Results: We identified 6 nonsynonymous heterozygous mutations (5 novel and 1 recurrent), 1 splice site mutation, and 1 deletion of 10 amino acids (not accounted in the mutant frequency) in 11 unrelated patients, accounting for a mutant frequency of 5.6% (1/18) in FALS, 2.3% (8/353) in SALS, and 8.3% (1/12) in ALS-FTD. The deletion of 10 amino acids was detected in 1 clinically undetermined male with an ALS family history who had atrophy in hand muscles and myotonic discharges revealed by EMG. The novel p. P36R mutation was identified in 1 FALS index, 1 patient with SALS, and 1 ALS-FTD. The splicing mutation (c.174-2A>G) caused in-frame skipping of the entire exon 6. The rest missense mutations including p.D40G, p.V128M, p.S229R, p.R302C and p.G491R were found in 6 unrelated patients with SALS.
Conclusions: The ANXA11 gene is one of the most frequently mutated genes in Chinese patients with SALS. A canonical splice site mutation leading to skipping of the entire exon 6 further supports the loss-of-function mechanism. In addition, the study findings further expand the ANXA11 phenotype, first highlighting its pathogenic role in ALS-FTD.
Figures
References
-
- Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol 2011;7:603–615. - PubMed
-
- Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med 2017;377:162–172. - PubMed
-
- Brooks BR, Miller RG, Swash M, Munsat TL; World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000;1:293–299. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
