Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models
- PMID: 29846515
- PMCID: PMC6067127
- DOI: 10.1093/carcin/bgy073
Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models
Abstract
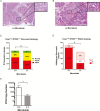
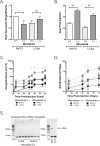
Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer death in the United States yet data are scant regarding host factors influencing pancreatic carcinogenesis. Increasing evidence support the role of the host microbiota in carcinogenesis but its role in PDAC is not well established. Herein, we report that antibiotic-mediated microbial depletion of KrasG12D/PTENlox/+ mice showed a decreased proportion of poorly differentiated tumors compared to microbiota-intact KrasG12D/PTENlox/+ mice. Subsequent 16S rRNA PCR showed that ~50% of KrasG12D/PTENlox/+ mice with PDAC harbored intrapancreatic bacteria. To determine if a similar observation in humans correlates with presence of PDAC, benign and malignant human pancreatic surgical specimens demonstrated a microbiota by 16S bacterial sequencing and culture confirmation. However, the microbial composition did not differentiate PDAC from non-PDAC tissue. Furthermore, murine pancreas did not naturally acquire a pancreatic microbiota, as germ-free mice transferred to specific pathogen-free housing failed to acquire intrapancreatic bacteria over time, which was not augmented by a murine model of colitis. Finally, antibiotic-mediated microbial depletion of Nod-SCID mice, compared to microbiota-intact, showed increased time to PDAC xenograft formation, smaller tumors, and attenuated growth. Interestingly, both xenograft cohorts were devoid of intratumoral bacteria by 16S rRNA PCR, suggesting that intrapancreatic/intratumoral microbiota is not the sole driver of PDAC acceleration. Xenografts from microbiota-intact mice demonstrated innate immune suppression by immunohistochemistry and differential regulation of oncogenic pathways as determined by RNA sequencing. Our work supports a long-distance role of the intestinal microbiota on PDAC progression and opens new research avenues regarding pancreatic carcinogenesis.
© The Author(s) 2018. Published by Oxford University Press.
Figures



References
-
- Hruban R.H., et al. (2000)Progression model for pancreatic cancer. Clin. Cancer Res., 6, 2969–2972. - PubMed
-
- Bailey P., et al. ; Pancreatic Cancer Genome Initiative. (2016)Genomic analyses identify molecular subtypes of pancreatic cancer. Nature, 531, 47–52. - PubMed
-
- Brat D.J., et al. (1998)Progression of pancreatic intraductal neoplasias to infiltrating adenocarcinoma of the pancreas. Am. J. Surg. Pathol., 22, 163–169. - PubMed
-
- Hruban R.H., et al. (2001)Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am. J. Surg. Pathol., 25, 579–586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

