Clostridium difficile: Investigating Transmission Patterns Between Infected and Colonized Patients Using Whole Genome Sequencing
- PMID: 29846557
- PMCID: PMC6321846
- DOI: 10.1093/cid/ciy457
Clostridium difficile: Investigating Transmission Patterns Between Infected and Colonized Patients Using Whole Genome Sequencing
Abstract
Background: Whole genome sequencing (WGS) studies can enhance our understanding of the role of patients with asymptomatic Clostridium difficile colonization in transmission.
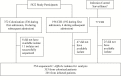
Methods: Isolates obtained from patients with Clostridium difficile infection (CDI) and colonization identified in a study conducted during 2006-2007 at 6 Canadian hospitals underwent typing by pulsed-field gel electrophoresis, multilocus sequence typing, and WGS. Isolates from incident CDI cases not in the initial study were also sequenced where possible. Ward movement and typing data were combined to identify plausible donors for each CDI case, as defined by shared time and space within predefined limits. Proportions of plausible donors for CDI cases that were colonized, infected, or both were examined.
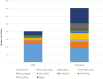
Results: Five hundred fifty-four isolates were sequenced successfully, 353 from colonized patients and 201 from CDI cases. The NAP1/027/ST1 strain was the most common strain, found in 124 (62%) of infected and 92 (26%) of colonized patients. A donor with a plausible ward link was found for 81 CDI cases (40%) using WGS with a threshold of ≤2 single nucleotide polymorphisms to determine relatedness. Sixty-five (32%) CDI cases could be linked to both infected and colonized donors. Exclusive linkages to infected and colonized donors were found for 28 (14%) and 12 (6%) CDI cases, respectively.
Conclusions: Colonized patients contribute to transmission, but CDI cases are more likely linked to other infected patients than colonized patients in this cohort with high rates of the NAP1/027/ST1 strain, highlighting the importance of local prevalence of virulent strains in determining transmission dynamics.
Figures
Comment in
-
The Challenges of Tracking Clostridium difficile to Its Source in Hospitalized Patients.Clin Infect Dis. 2019 Jan 7;68(2):210-212. doi: 10.1093/cid/ciy461. Clin Infect Dis. 2019. PMID: 29846537 Free PMC article. No abstract available.
References
-
- Evans CT, Safdar N. Current trends in the epidemiology and outcomes of Clostridium difficile infection. Clin Infect Dis 2015; 60(Suppl 2):S66–71. - PubMed
-
- Martin JS, Monaghan TM, Wilcox MH. Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nat Rev Gastroenterol Hepatol 2016; 13:206–16. - PubMed
-
- Cohen SH, Gerding DN, Johnson S, et al. ; Society for Healthcare Epidemiology of America; Infectious Diseases Society of America Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 2010; 31:431–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

