The archaeal ATPase PINA interacts with the helicase Hjm via its carboxyl terminal KH domain remodeling and processing replication fork and Holliday junction
- PMID: 29846688
- PMCID: PMC6061704
- DOI: 10.1093/nar/gky451
The archaeal ATPase PINA interacts with the helicase Hjm via its carboxyl terminal KH domain remodeling and processing replication fork and Holliday junction
Abstract
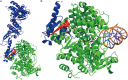
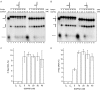
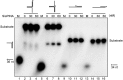
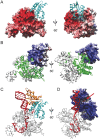
PINA is a novel ATPase and DNA helicase highly conserved in Archaea, the third domain of life. The PINA from Sulfolobus islandicus (SisPINA) forms a hexameric ring in crystal and solution. The protein is able to promote Holliday junction (HJ) migration and physically and functionally interacts with Hjc, the HJ specific endonuclease. Here, we show that SisPINA has direct physical interaction with Hjm (Hel308a), a helicase presumably targeting replication forks. In vitro biochemical analysis revealed that Hjm, Hjc, and SisPINA are able to coordinate HJ migration and cleavage in a concerted way. Deletion of the carboxyl 13 amino acid residues impaired the interaction between SisPINA and Hjm. Crystal structure analysis showed that the carboxyl 70 amino acid residues fold into a type II KH domain which, in other proteins, functions in binding RNA or ssDNA. The KH domain not only mediates the interactions of PINA with Hjm and Hjc but also regulates the hexameric assembly of PINA. Our results collectively suggest that SisPINA, Hjm and Hjc work together to function in replication fork regression, HJ formation and HJ cleavage.
Figures






Similar articles
-
Genetic Study of Four Candidate Holliday Junction Processing Proteins in the Thermophilic Crenarchaeon Sulfolobus acidocaldarius.Int J Mol Sci. 2022 Jan 9;23(2):707. doi: 10.3390/ijms23020707. Int J Mol Sci. 2022. PMID: 35054893 Free PMC article.
-
Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.J Mol Biol. 2017 Apr 7;429(7):1009-1029. doi: 10.1016/j.jmb.2017.02.016. Epub 2017 Feb 24. J Mol Biol. 2017. PMID: 28238763 Free PMC article.
-
Dissection of the functional domains of an archaeal Holliday junction helicase.DNA Repair (Amst). 2012 Feb 1;11(2):102-11. doi: 10.1016/j.dnarep.2011.10.009. Epub 2011 Nov 6. DNA Repair (Amst). 2012. PMID: 22062475
-
Molecular machines in archaeal DNA replication.Curr Opin Chem Biol. 2011 Oct;15(5):614-9. doi: 10.1016/j.cbpa.2011.07.017. Epub 2011 Aug 16. Curr Opin Chem Biol. 2011. PMID: 21852183 Review.
-
DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication forks in Escherichia coli.Genes (Basel). 2020 Apr 26;11(5):471. doi: 10.3390/genes11050471. Genes (Basel). 2020. PMID: 32357475 Free PMC article. Review.
Cited by
-
Phylogenetic Diversity of Lhr Proteins and Biochemical Activities of the Thermococcales aLhr2 DNA/RNA Helicase.Biomolecules. 2021 Jun 26;11(7):950. doi: 10.3390/biom11070950. Biomolecules. 2021. PMID: 34206878 Free PMC article.
-
The KH domain facilitates the substrate specificity and unwinding processivity of DDX43 helicase.J Biol Chem. 2021 Jan-Jun;296:100085. doi: 10.1074/jbc.RA120.015824. Epub 2020 Nov 23. J Biol Chem. 2021. PMID: 33199368 Free PMC article.
-
Archaeal Hel308 suppresses recombination through a catalytic switch that controls DNA annealing.Nucleic Acids Res. 2023 Sep 8;51(16):8563-8574. doi: 10.1093/nar/gkad572. Nucleic Acids Res. 2023. PMID: 37409572 Free PMC article.
-
Phosphorylation of the Archaeal Holliday Junction Resolvase Hjc Inhibits Its Catalytic Activity and Facilitates DNA Repair in Sulfolobus islandicus REY15A.Front Microbiol. 2019 May 31;10:1214. doi: 10.3389/fmicb.2019.01214. eCollection 2019. Front Microbiol. 2019. PMID: 31214148 Free PMC article.
-
Genetic Study of Four Candidate Holliday Junction Processing Proteins in the Thermophilic Crenarchaeon Sulfolobus acidocaldarius.Int J Mol Sci. 2022 Jan 9;23(2):707. doi: 10.3390/ijms23020707. Int J Mol Sci. 2022. PMID: 35054893 Free PMC article.
References
-
- Branzei D., Foiani M.. Maintaining genome stability at the replication fork. Nat. Rev. 2010; 11:208–219. - PubMed
-
- Friedberg E.C., Walker G.C., Siede W.. DNA Repair and Mutagenesis. 2005; Washington, D.C: ASM Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

