Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer
- PMID: 29848291
- PMCID: PMC5977541
- DOI: 10.1186/s12885-018-4448-9
Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer
Abstract
Background: Gene-expression companion diagnostic tests, such as the Oncotype DX test, assess the risk of early stage Estrogen receptor (ER) positive (+) breast cancers, and guide clinicians in the decision of whether or not to use chemotherapy. However, these tests are typically expensive, time consuming, and tissue-destructive.
Methods: In this paper, we evaluate the ability of computer-extracted nuclear morphology features from routine hematoxylin and eosin (H&E) stained images of 178 early stage ER+ breast cancer patients to predict corresponding risk categories derived using the Oncotype DX test. A total of 216 features corresponding to the nuclear shape and architecture categories from each of the pathologic images were extracted and four feature selection schemes: Ranksum, Principal Component Analysis with Variable Importance on Projection (PCA-VIP), Maximum-Relevance, Minimum Redundancy Mutual Information Difference (MRMR MID), and Maximum-Relevance, Minimum Redundancy - Mutual Information Quotient (MRMR MIQ), were employed to identify the most discriminating features. These features were employed to train 4 machine learning classifiers: Random Forest, Neural Network, Support Vector Machine, and Linear Discriminant Analysis, via 3-fold cross validation.
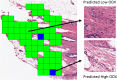
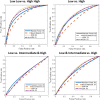
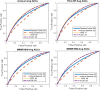
Results: The four sets of risk categories, and the top Area Under the receiver operating characteristic Curve (AUC) machine classifier performances were: 1) Low ODx and Low mBR grade vs. High ODx and High mBR grade (Low-Low vs. High-High) (AUC = 0.83), 2) Low ODx vs. High ODx (AUC = 0.72), 3) Low ODx vs. Intermediate and High ODx (AUC = 0.58), and 4) Low and Intermediate ODx vs. High ODx (AUC = 0.65). Trained models were tested independent validation set of 53 cases which comprised of Low and High ODx risk, and demonstrated per-patient accuracies ranging from 75 to 86%.
Conclusion: Our results suggest that computerized image analysis of digitized H&E pathology images of early stage ER+ breast cancer might be able predict the corresponding Oncotype DX risk categories.
Conflict of interest statement
Ethics approval and consent to participate
The study was HIPAA compliant and was approved by the Institutional Review Board at the University Hospitals Case Medical Center. The informed consent was waived by the institutional review board for this retrospective study.
Competing interests
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. He is also a scientific advisory consultant for Inspirata Inc. In addition, he currently serves as a scientific advisory board member for Inspirata Inc. and for Astrazeneca. He also has sponsored research agreements with Philips and Inspirata Inc. His technology has been licensed to Elucid Bioimaging and Inspirata Inc. He is also involved in a NIH U24 grant with PathCore Inc. and a R01 with Inspirata Inc. Drs John Tomaszewski. Michael Feldman and Shridar Ganesan are members of the scientific advisory board of Inspirata, Inc. a digital pathology start-up company, and receives board fees and stock options. The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. - DOI - PubMed
-
- Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner FL, Watson D, Bryant J, Costantino JP, Geyer CE, Wickerham DL, Wolmark N. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast Cancer. J Clin Oncol. 2006;24(23):3726–3734. doi: 10.1200/JCO.2005.04.7985. - DOI - PubMed
-
- Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Dees EC, Perez EA, Olson JA, Zujewski J, Lively T, Badve SS, Saphner TJ, Wagner LI, Whelan TJ, Ellis MJ, Paik S, Wood WC, Ravdin P, Keane MM, Gomez Moreno HL, Reddy PS, Goggins TF, Mayer IA, Brufsky AM, Toppmeyer DL, Kaklamani VG, Atkins JN, Berenberg JL, Sledge GW. Prospective validation of a 21-gene expression assay in breast Cancer. N Engl J Med. 2015;373(21):2005–2014. doi: 10.1056/NEJMoa1510764. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA216579/CA/NCI NIH HHS/United States
- 1U24CA199374-01/CA/NCI NIH HHS/United States
- U24 CA199374/CA/NCI NIH HHS/United States
- R21CA179327-01/CA/NCI NIH HHS/United States
- R01DK098503-02/DK/NIDDK NIH HHS/United States
- T32 DK007470/DK/NIDDK NIH HHS/United States
- R21 CA195152/CA/NCI NIH HHS/United States
- R01 DK098503/DK/NIDDK NIH HHS/United States
- R01 CA202752/CA/NCI NIH HHS/United States
- R01 CA208236/CA/NCI NIH HHS/United States
- LC130463/U.S. Department of Defense
- C06 RR012463/RR/NCRR NIH HHS/United States
- VelaSano/Cleveland Clinic
- Wallace Coulter Foundation/Case Western Reserve University Department of Biomedical Engineering
- Comprehensive Cancer Center Pilot Grant/School of Medicine, Case Western Reserve University
- R01CA202752-01A1/CA/NCI NIH HHS/United States
- R21CA195152-01/CA/NCI NIH HHS/United States
- PC120857/U.S. Department of Defense
- R21 CA179327/CA/NCI NIH HHS/United States
- Prostate Cancer Idea Development Award/U.S. Department of Defense
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

