Genetic and epidemiological analysis of norovirus from children with gastroenteritis in Botswana, 2013-2015
- PMID: 29848303
- PMCID: PMC5975618
- DOI: 10.1186/s12879-018-3157-y
Genetic and epidemiological analysis of norovirus from children with gastroenteritis in Botswana, 2013-2015
Abstract
Background: Norovirus is a leading cause of viral gastroenteritis worldwide with a peak of disease seen in children. The epidemiological analysis regarding the virus strains in Africa is limited. The first report of norovirus in Botswana was in 2010 and currently, the prevalence and circulating genotypes of norovirus are unknown, as the country has no systems to report the norovirus cases. This study investigated the prevalence, patterns and molecular characteristics of norovirus infections among children ≤5 years of age admitted with acute gastroenteritis at four hospitals in Botswana.
Methods: A total of 484 faecal samples were collected from children who were admitted with acute gastroenteritis during the rotavirus vaccine impact survey between July 2013 and December 2015. Norovirus was detected using real-time RT-PCR. Positive samples were genotyped using conventional RT-PCR followed by partial sequencing of the capsid and RdRp genes. Norovirus strains were determined by nucleotide sequence analysis using the online Norovirus Genotyping Tool Version 1.0, and confirmed using maximum likelihood tree construction as implemented in MEGA 6.0.
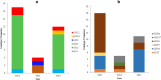
Results: The prevalence of norovirus was 9.3% (95% CI 6.7-11.9). The genotype diversity was dominated by the GII.4 strain at 69.7%. This was followed by GII.2, GII.12 each at 9.1%, GI.9 at 6.6% and GII.6, GII.10 each at 3.0%. The most common combined RdRp/Capsid genotype was the GII.Pe/GII.4 Sydney 2012. Norovirus was detected during most part of the year; however, there was a preponderance of cases in the wet season (December to March).
Conclusion: The study showed a possible decline of norovirus infections in the last 10 years since the first report. An upward trend seen between 2013 and 2015 may be attributable to the success of rotavirus vaccine introductions in 2012. Knowledge of circulating genotypes, seasonal trends and overall prevalence is critical for prevention programming and possible future vaccine design implications.
Keywords: Botswana; GII.4 variants; GII.Pe-GII.4; Genotyping; Norovirus.
Conflict of interest statement
Ethics approval and consent to participate
Written informed consent was obtained from all parents and guardians of the children that were enrolled in this study. Ethical approval for the study, to perform molecular analyses on the samples was obtained from Botswana’s institutional review board, the Health Research and Development Council (HRDC). Furthermore, permission to collect data, samples from patients and subsequent analysis was sought from HRDC, McMaster University and University of Pennsylvania as reported in the previous study [22]. This study was conducted according to the principles expressed in the Declaration of Helsinki.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Molecular epidemiology and genetic diversity of norovirus among hospitalized children with acute gastroenteritis in Tianjin, China, 2018-2020.BMC Infect Dis. 2021 Jul 14;21(1):682. doi: 10.1186/s12879-021-06375-2. BMC Infect Dis. 2021. PMID: 34261441 Free PMC article.
-
Clinical and epidemiological characteristics of norovirus gastroenteritis among hospitalized children in Lebanon.World J Gastroenterol. 2016 Dec 28;22(48):10557-10565. doi: 10.3748/wjg.v22.i48.10557. World J Gastroenterol. 2016. PMID: 28082807 Free PMC article.
-
Prevalence and genetic diversity of norovirus genogroup II in children less than 5 years of age with acute gastroenteritis in Tehran, Iran.Med Microbiol Immunol. 2018 Aug;207(3-4):201-210. doi: 10.1007/s00430-018-0541-6. Epub 2018 Apr 4. Med Microbiol Immunol. 2018. PMID: 29619604
-
Norovirus Infections and Disease in Lower-MiddleandLow-Income Countries, 1997⁻2018.Viruses. 2019 Apr 10;11(4):341. doi: 10.3390/v11040341. Viruses. 2019. PMID: 30974898 Free PMC article. Review.
-
Global prevalence and genotype distribution of norovirus infection in children with gastroenteritis: A meta-analysis on 6 years of research from 2015 to 2020.Rev Med Virol. 2022 Jan;32(1):e2237. doi: 10.1002/rmv.2237. Epub 2021 Apr 1. Rev Med Virol. 2022. PMID: 33793023 Review.
Cited by
-
Norovirus: Facts and Reflections from Past, Present, and Future.Viruses. 2021 Nov 30;13(12):2399. doi: 10.3390/v13122399. Viruses. 2021. PMID: 34960668 Free PMC article. Review.
-
Global prevalence of norovirus gastroenteritis after emergence of the GII.4 Sydney 2012 variant: a systematic review and meta-analysis.Front Public Health. 2024 Jun 27;12:1373322. doi: 10.3389/fpubh.2024.1373322. eCollection 2024. Front Public Health. 2024. PMID: 38993708 Free PMC article.
-
Prevalence and associated factors of norovirus infections among patients with diarrhea in the Amhara national regional state, Ethiopia.BMC Infect Dis. 2024 Sep 27;24(1):1053. doi: 10.1186/s12879-024-09988-5. BMC Infect Dis. 2024. PMID: 39333942 Free PMC article.
-
Genetic Diversity of Norovirus in Children with Acute Gastroenteritis in Southwest Nigeria, 2015-2017.Viruses. 2023 Feb 28;15(3):644. doi: 10.3390/v15030644. Viruses. 2023. PMID: 36992354 Free PMC article.
-
Epidemiological characteristics and genetic diversity of norovirus infections among outpatient children with diarrhea under 5 years of age in Beijing, China, 2011-2018.Gut Pathog. 2021 Dec 24;13(1):77. doi: 10.1186/s13099-021-00473-x. Gut Pathog. 2021. PMID: 34952625 Free PMC article.
References
-
- Vicentini F, Denadai W, Gomes YM, Rose TL, Ferreira MS, Le Moullac-Vaidye B, Le Pendu J, Leite JP, Miagostovich MP, Spano LC. Molecular characterization of noroviruses and HBGA from infected Quilombola children in Espirito Santo state, Brazil. PLoS One. 2013;8(7):e69348. doi: 10.1371/journal.pone.0069348. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

