Bayesian Networks Predict Neuronal Transdifferentiation
- PMID: 29848620
- PMCID: PMC6027867
- DOI: 10.1534/g3.118.200401
Bayesian Networks Predict Neuronal Transdifferentiation
Abstract
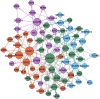
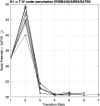
We employ the language of Bayesian networks to systematically construct gene-regulation topologies from deep-sequencing single-nucleus RNA-Seq data for human neurons. From the perspective of the cell-state potential landscape, we identify attractors that correspond closely to different neuron subtypes. Attractors are also recovered for cell states from an independent data set confirming our models accurate description of global genetic regulations across differing cell types of the neocortex (not included in the training data). Our model recovers experimentally confirmed genetic regulations and community analysis reveals genetic associations in common pathways. Via a comprehensive scan of all theoretical three-gene perturbations of gene knockout and overexpression, we discover novel neuronal trans-differrentiation recipes (including perturbations of SATB2, GAD1, POU6F2 and ADARB2) for excitatory projection neuron and inhibitory interneuron subtypes.
Keywords: gene regulation network; neuroscience; systems biology.
Copyright © 2018 Ainsworth et al.
Figures
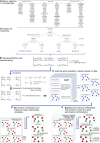

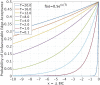
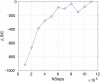
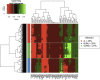

References
-
- Adams B. D., Claffey K. P., White B. A., 2009. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology 150: 14–23. 10.1210/en.2008-0984 - DOI - PMC - PubMed
-
- Blondel V. D., Guillaume J.-L., Lambiotte R., Lefebvre E., 2008. Fast unfolding of communities in large networks. J. Stat. Mech. 2008: P10008 10.1088/1742-5468/2008/10/P10008 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
