MODE-TASK: large-scale protein motion tools
- PMID: 29850770
- PMCID: PMC6198866
- DOI: 10.1093/bioinformatics/bty427
MODE-TASK: large-scale protein motion tools
Abstract
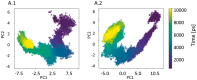
Summary: MODE-TASK, a novel and versatile software suite, comprises Principal Component Analysis, Multidimensional Scaling, and t-Distributed Stochastic Neighbor Embedding techniques using Molecular Dynamics trajectories. MODE-TASK also includes a Normal Mode Analysis tool based on Anisotropic Network Model so as to provide a variety of ways to analyse and compare large-scale motions of protein complexes for which long MD simulations are prohibitive. Beside the command line function, a GUI has been developed as a PyMOL plugin.
Availability and implementation: MODE-TASK is open source, and available for download from https://github.com/RUBi-ZA/MODE-TASK. It is implemented in Python and C++. It is compatible with Python 2.x and Python 3.x and can be installed by Conda.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
Similar articles
-
MD-TASK: a software suite for analyzing molecular dynamics trajectories.Bioinformatics. 2017 Sep 1;33(17):2768-2771. doi: 10.1093/bioinformatics/btx349. Bioinformatics. 2017. PMID: 28575169 Free PMC article.
-
Epock: rapid analysis of protein pocket dynamics.Bioinformatics. 2015 May 1;31(9):1478-80. doi: 10.1093/bioinformatics/btu822. Epub 2014 Dec 12. Bioinformatics. 2015. PMID: 25505095 Free PMC article.
-
MD DaVis: interactive data visualization of protein molecular dynamics.Bioinformatics. 2022 Jun 13;38(12):3299-3301. doi: 10.1093/bioinformatics/btac314. Bioinformatics. 2022. PMID: 35532115
-
MDBuilder: a PyMOL plugin for the preparation of molecular dynamics simulations.Brief Bioinform. 2023 Mar 19;24(2):bbad057. doi: 10.1093/bib/bbad057. Brief Bioinform. 2023. PMID: 36790845
-
The National Rehabilitation Information Center (NARIC).J Med Libr Assoc. 2022 Jul 1;110(3):388-391. doi: 10.5195/jmla.2022.1515. J Med Libr Assoc. 2022. PMID: 36589293 Free PMC article. Review.
Cited by
-
Protein dynamics developments for the large scale and cryoEM: case study of ProDy 2.0.Acta Crystallogr D Struct Biol. 2022 Apr 1;78(Pt 4):399-409. doi: 10.1107/S2059798322001966. Epub 2022 Mar 16. Acta Crystallogr D Struct Biol. 2022. PMID: 35362464 Free PMC article. Review.
-
Protein Function Analysis through Machine Learning.Biomolecules. 2022 Sep 6;12(9):1246. doi: 10.3390/biom12091246. Biomolecules. 2022. PMID: 36139085 Free PMC article. Review.
-
A Novel Therapeutic Peptide Blocks SARS-CoV-2 Spike Protein Binding with Host Cell ACE2 Receptor.Drugs R D. 2021 Sep;21(3):273-283. doi: 10.1007/s40268-021-00357-0. Epub 2021 Jul 29. Drugs R D. 2021. PMID: 34324175 Free PMC article.
-
Collective variable discovery in the age of machine learning: reality, hype and everything in between.RSC Adv. 2022 Sep 2;12(38):25010-25024. doi: 10.1039/d2ra03660f. eCollection 2022 Aug 30. RSC Adv. 2022. PMID: 36199882 Free PMC article. Review.
-
Side-by-side comparison of Notch- and C83 binding to γ-secretase in a complete membrane model at physiological temperature.RSC Adv. 2020 Aug 24;10(52):31215-31232. doi: 10.1039/d0ra04683c. eCollection 2020 Aug 21. RSC Adv. 2020. PMID: 35520661 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources