Phylogenomic Analysis of Lactobacillus curvatus Reveals Two Lineages Distinguished by Genes for Fermenting Plant-Derived Carbohydrates
- PMID: 29850855
- PMCID: PMC6007345
- DOI: 10.1093/gbe/evy106
Phylogenomic Analysis of Lactobacillus curvatus Reveals Two Lineages Distinguished by Genes for Fermenting Plant-Derived Carbohydrates
Abstract
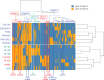
Lactobacillus curvatus is a lactic acid bacterium encountered in many different types of fermented food (meat, seafood, vegetables, and cereals). Although this species plays an important role in the preservation of these foods, few attempts have been made to assess its genomic diversity. This study uses comparative analyses of 13 published genomes (complete or draft) to better understand the evolutionary processes acting on the genome of this species. Phylogenomic analysis, based on a coalescent model of evolution, revealed that the 6,742 sites of single nucleotide polymorphism within the L. curvatus core genome delineate two major groups, with lineage 1 represented by the newly sequenced strain FLEC03, and lineage 2 represented by the type-strain DSM20019. The two lineages could also be distinguished by the content of their accessory genome, which sheds light on a long-term evolutionary process of lineage-dependent genetic acquisition and the possibility of population structure. Interestingly, one clade from lineage 2 shared more accessory genes with strains of lineage 1 than with other strains of lineage 2, indicating recent convergence in carbohydrate catabolism. Both lineages had a wide repertoire of accessory genes involved in the fermentation of plant-derived carbohydrates that are released from polymers of α/β-glucans, α/β-fructans, and N-acetylglucosan. Other gene clusters were distributed among strains according to the type of food from which the strains were isolated. These results give new insight into the ecological niches in which L. curvatus may naturally thrive (such as silage or compost heaps) in addition to fermented food.
Figures



Similar articles
-
Comparative genomics of Lactobacillus curvatus enables prediction of traits relating to adaptation and strategies of assertiveness in sausage fermentation.Int J Food Microbiol. 2018 Dec 2;286:37-47. doi: 10.1016/j.ijfoodmicro.2018.06.025. Epub 2018 Jul 4. Int J Food Microbiol. 2018. PMID: 30031987
-
Detection and genomic characterization of motility in Lactobacillus curvatus: confirmation of motility in a species outside the Lactobacillus salivarius clade.Appl Environ Microbiol. 2015 Feb;81(4):1297-1308. doi: 10.1128/AEM.03594-14. Appl Environ Microbiol. 2015. PMID: 25501479 Free PMC article.
-
Genetic screening of Lactobacillus sakei and Lactobacillus curvatus strains for their peptidolytic system and amino acid metabolism, and comparison of their volatilomes in a model system.Syst Appl Microbiol. 2011 Jul;34(5):311-20. doi: 10.1016/j.syapm.2010.12.006. Epub 2011 May 12. Syst Appl Microbiol. 2011. PMID: 21570226
-
Functional genomics of probiotic Lactobacilli.J Clin Gastroenterol. 2008 Sep;42 Suppl 3 Pt 2:S160-2. doi: 10.1097/MCG.0b013e31817da140. J Clin Gastroenterol. 2008. PMID: 18685516 Review.
-
Probiotic characters of Bifidobacterium and Lactobacillus are a result of the ongoing gene acquisition and genome minimization evolutionary trends.Microb Pathog. 2017 Oct;111:118-131. doi: 10.1016/j.micpath.2017.08.021. Epub 2017 Aug 18. Microb Pathog. 2017. PMID: 28826768 Review.
Cited by
-
Effects of Lactobacillus curvatus MG5246 on inflammatory markers in Porphyromonas gingivalis lipopolysaccharide-sensitized human gingival fibroblasts and periodontitis rat model.Food Sci Biotechnol. 2021 Nov 26;31(1):111-120. doi: 10.1007/s10068-021-01009-4. eCollection 2022 Jan. Food Sci Biotechnol. 2021. PMID: 35059235 Free PMC article.
-
Latilactobacillus curvatus: A Candidate Probiotic with Excellent Fermentation Properties and Health Benefits.Foods. 2020 Sep 25;9(10):1366. doi: 10.3390/foods9101366. Foods. 2020. PMID: 32993033 Free PMC article. Review.
-
Increase in Lactulose Content in a Hot-Alkaline-Based System through Fermentation with a Selected Lactic Acid Bacteria Strain Followed by the β-Galactosidase Catalysis Process.Foods. 2023 Nov 29;12(23):4317. doi: 10.3390/foods12234317. Foods. 2023. PMID: 38231767 Free PMC article.
-
Holistic integration of omics data reveals the drivers that shape the ecology of microbial meat spoilage scenarios.Front Microbiol. 2023 Oct 18;14:1286661. doi: 10.3389/fmicb.2023.1286661. eCollection 2023. Front Microbiol. 2023. PMID: 37920261 Free PMC article.
-
New Insight into Bacterial Interaction with the Matrix of Plant-Based Fermented Foods.Foods. 2021 Jul 10;10(7):1603. doi: 10.3390/foods10071603. Foods. 2021. PMID: 34359473 Free PMC article. Review.
References
-
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC.. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1,2-propanediol degradation. J Bacteriol. 181(19):5967–5975. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

