Anti-osteogenic function of a LIM-homeodomain transcription factor LMX1B is essential to early patterning of the calvaria
- PMID: 29852132
- PMCID: PMC6197925
- DOI: 10.1016/j.ydbio.2018.05.022
Anti-osteogenic function of a LIM-homeodomain transcription factor LMX1B is essential to early patterning of the calvaria
Abstract
The calvaria (upper part of the skull) is made of plates of bone and fibrous joints (sutures and fontanelles), and the proper balance and organization of these components are crucial to normal development of the calvaria. In a mouse embryo, the calvaria develops from a layer of head mesenchyme that surrounds the brain from shortly after mid-gestation. The mesenchyme just above the eye (supra-orbital mesenchyme, SOM) generates ossification centers for the bones, which then grow toward the apex gradually. In contrast, the mesenchyme apical to SOM (early migrating mesenchyme, EMM), including the area at the vertex, does not generate an ossification center. As a result, the dorsal midline of the head is occupied by sutures and fontanelles at birth. To date, the molecular basis for this regional difference in developmental programs is unknown. The current study provides vital insights into the genetic regulation of calvarial patterning. First, we showed that osteogenic signals were active in both EMM and SOM during normal development, which suggested the presence of an anti-osteogenic factor in EMM to counter the effect of these signals. Subsequently, we identified Lmx1b as an anti-osteogenic gene that was expressed in EMM but not in SOM. Furthermore, head mesenchyme-specific deletion of Lmx1b resulted in heterotopic ossification from EMM at the vertex, and craniosynostosis affecting multiple sutures. Conversely, forced expression of Lmx1b in SOM was sufficient to inhibit osteogenic specification. Therefore, we conclude that Lmx1b plays a key role as an anti-osteogenic factor in patterning the head mesenchyme into areas with different osteogenic competence. In turn, this patterning event is crucial to generating the proper organization of the bones and soft tissue joints of the calvaria.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures



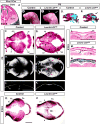



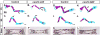
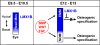
Similar articles
-
The limb dorsoventral axis: Lmx1b's role in development, pathology, evolution, and regeneration.Dev Dyn. 2024 Sep;253(9):798-814. doi: 10.1002/dvdy.695. Epub 2024 Jan 30. Dev Dyn. 2024. PMID: 38288855 Free PMC article. Review.
-
Molecular patterning of the embryonic cranial mesenchyme revealed by genome-wide transcriptional profiling.Dev Biol. 2019 Nov 15;455(2):434-448. doi: 10.1016/j.ydbio.2019.07.015. Epub 2019 Jul 24. Dev Biol. 2019. PMID: 31351040 Free PMC article.
-
Lineage-specific mutation of Lmx1b provides new insights into distinct regulation of suture development in different areas of the calvaria.Front Physiol. 2023 Aug 1;14:1225118. doi: 10.3389/fphys.2023.1225118. eCollection 2023. Front Physiol. 2023. PMID: 37593235 Free PMC article.
-
Multiple calvarial defects in lmx1b mutant mice.Dev Genet. 1998;22(4):314-20. doi: 10.1002/(SICI)1520-6408(1998)22:4<314::AID-DVG2>3.0.CO;2-9. Dev Genet. 1998. PMID: 9664684
-
The Development of the Calvarial Bones and Sutures and the Pathophysiology of Craniosynostosis.Curr Top Dev Biol. 2015;115:131-56. doi: 10.1016/bs.ctdb.2015.07.004. Epub 2015 Oct 1. Curr Top Dev Biol. 2015. PMID: 26589924 Review.
Cited by
-
En1 and Lmx1b do not recapitulate embryonic dorsal-ventral limb patterning functions during mouse digit tip regeneration.Cell Rep. 2022 Nov 22;41(8):111701. doi: 10.1016/j.celrep.2022.111701. Cell Rep. 2022. PMID: 36417876 Free PMC article.
-
LIM Homeobox Transcription Factor 1-β Expression is Upregulated in Patients with Osteolysis after Total Ankle Arthroplasty and Inhibits Receptor Activator of Nuclear Factor-κB Ligand-Induced Osteoclast Differentiation in Vitro.J Bone Metab. 2022 Aug;29(3):165-174. doi: 10.11005/jbm.2022.29.3.165. Epub 2022 Aug 31. J Bone Metab. 2022. PMID: 36153852 Free PMC article.
-
The limb dorsoventral axis: Lmx1b's role in development, pathology, evolution, and regeneration.Dev Dyn. 2024 Sep;253(9):798-814. doi: 10.1002/dvdy.695. Epub 2024 Jan 30. Dev Dyn. 2024. PMID: 38288855 Free PMC article. Review.
-
Molecular patterning of the embryonic cranial mesenchyme revealed by genome-wide transcriptional profiling.Dev Biol. 2019 Nov 15;455(2):434-448. doi: 10.1016/j.ydbio.2019.07.015. Epub 2019 Jul 24. Dev Biol. 2019. PMID: 31351040 Free PMC article.
-
A tale of two cities: The genetic mechanisms governing calvarial bone development.Genesis. 2019 Jan;57(1):e23248. doi: 10.1002/dvg.23248. Epub 2018 Oct 3. Genesis. 2019. PMID: 30155972 Free PMC article. Review.
References
-
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A Twist Code Determines the Onset of Osteoblast Differentiation. Developmental Cell. 2004;6:423–435. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Chen H, Ovchinnikov D, Pressman CL, Aulehla A, Lun Y, Johnson RL. Multiple calvarial defects inlmx1b mutant mice. Developmental Genetics. 1998a;22:314–320. - PubMed
-
- Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg KC, Pepicelli CV, Gan L, Lee B, Johnson RL. Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat Genet. 1998b;19:51–55. - PubMed
-
- Chen L, Li D, Li C, Engel A, Deng C-X. A Ser250Trp substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone. 2003;33:169–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

