Predicting Interactions between Virus and Host Proteins Using Repeat Patterns and Composition of Amino Acids
- PMID: 29854357
- PMCID: PMC5966669
- DOI: 10.1155/2018/1391265
Predicting Interactions between Virus and Host Proteins Using Repeat Patterns and Composition of Amino Acids
Abstract
Previous methods for predicting protein-protein interactions (PPIs) were mainly focused on PPIs within a single species, but PPIs across different species have recently emerged as an important issue in some areas such as viral infection. The primary focus of this study is to predict PPIs between virus and its targeted host, which are involved in viral infection. We developed a general method that predicts interactions between virus and host proteins using the repeat patterns and composition of amino acids. In independent testing of the method with PPIs of new viruses and hosts, it showed a high performance comparable to the best performance of other methods for single virus-host PPIs. In comparison of our method with others using same datasets, our method outperformed the others. The repeat patterns and composition of amino acids are simple, yet powerful features for predicting virus-host PPIs. The method developed in this study will help in finding new virus-host PPIs for which little information is available.
Figures
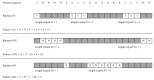

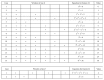

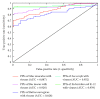
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

