A DNA-Encoded Library of Chemical Compounds Based on Common Scaffolding Structures Reveals the Impact of Ligand Geometry on Protein Recognition
- PMID: 29856130
- PMCID: PMC6126618
- DOI: 10.1002/cmdc.201800193
A DNA-Encoded Library of Chemical Compounds Based on Common Scaffolding Structures Reveals the Impact of Ligand Geometry on Protein Recognition
Abstract
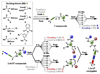
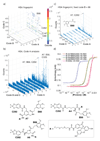
A DNA-encoded chemical library (DECL) with 1.2 million compounds was synthesized by combinatorial reaction of seven central scaffolds with two sets of 343×492 building blocks. Library screening by affinity capture revealed that for some target proteins, the chemical nature of building blocks dominated the selection results, whereas for other proteins, the central scaffold also crucially contributed to ligand affinity. Molecules based on a 3,5-bis(aminomethyl)benzoic acid core structure were found to bind human serum albumin with a Kd value of 6 nm, while compounds with the same substituents on an equidistant but flexible l-lysine scaffold showed 140-fold lower affinity. A 18 nm tankyrase-1 binder featured l-lysine as linking moiety, while molecules based on d-Lysine or (2S,4S)-amino-l-proline showed no detectable binding to the target. This work suggests that central scaffolds which predispose the orientation of chemical building blocks toward the protein target may enhance the screening productivity of encoded libraries.
Keywords: DNA-encoded chemical libraries; bifunctional ligands; human serum albumin; tankyrase-1.
© 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
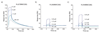

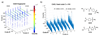
References
-
- Brenner S, Lerner RA. Proc Natl Acad Sci USA. 1992;89:5381–5383. - PMC - PubMed
- Arico-Muendel C, Zhu Z, Dickson H, Parks D, Keicher J, Deng J, Aquilani L, Coppo F, Graybill T, Lind K, Peat A, et al. Antimicrob Agents Chemother. 2015;59:3450–3459. - PMC - PubMed
- Winssinger N. Chimia (Aarau) 2013;67:340–348. - PubMed
- Blakskjaer P, Heitner T, Hansen NJ. Curr Opin Chem Biol. 2015;26:62–71. - PubMed
- Cuozzo JW, Centrella PA, Gikunju D, Habeshian S, Hupp CD, Keefe AD, Sigel EA, Soutter HH, Thomson HA, Zhang Y, Clark MA. Chembiochem. 2017;18:864–871. - PubMed
- Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J. Nat Chem. 2015;7:241–249. - PubMed
- Samain F, Ekblad T, Mikutis G, Zhong N, Zimmermann M, Nauer A, Bajic D, Decurtins W, Scheuermann J, Brown PJ, Hall J, et al. J Med Chem. 2015;58:5143–5149. - PubMed
- Leimbacher M, Zhang Y, Mannocci L, Stravs M, Geppert T, Scheuermann J, Schneider G, Neri D. Chemistry. 2012;18:7729–7737. - PubMed
- Franzini RM, Neri D, Scheuermann J. Acc Chem Res. 2014;47:1247–1255. - PubMed
- Salamon H, Klika Skopic M, Jung K, Bugain O, Brunschweiger A. ACS Chem Biol. 2016;11:296–307. - PubMed
- Neri D, Lerner R. Annu Rev Biochem. 2018;87:24. - PMC - PubMed
-
- Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, Nauer A, Zimmermann M, Samain F, Scheuermann J, Brown PJ, Hall J, et al. Angew Chem Int Ed Engl. 2015;54:3927–3931. - PubMed
-
- Satz AL. ACS Comb Sci. 2016;18:415–424. - PubMed
-
- Satz AL, Cai J, Chen Y, Goodnow R, Gruber F, Kowalczyk A, Petersen A, Naderi-Oboodi G, Orzechowski L, Strebel Q. Bioconjug Chem. 2015;26:1623–1632. - PubMed
- Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, et al. Nat Chem Biol. 2009;5:647–654. - PubMed
-
- Daguer JP, Zambaldo C, Ciobanu M, Morieux P, Barluenga S, Winssinger N. Chem Sci. 2015;6:739–744. - PMC - PubMed
- Deng H, Zhou J, Sundersingh FS, Summerfield J, Somers D, Messer JA, Satz AL, Ancellin N, Arico-Muendel CC, Sargent Bedard KL, Beljean A, et al. ACS Med Chem Lett. 2015;6:919–924. - PMC - PubMed
- Encinas L, O'Keefe H, Neu M, Remuinan MJ, Patel AM, Guardia A, Davie CP, Perez-Macias N, Yang H, Convery MA, Messer JA, et al. J Med Chem. 2014;57:1276–1288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

