Chemical cross-linking in the structural analysis of protein assemblies
- PMID: 29857191
- PMCID: PMC6051896
- DOI: 10.1016/j.ymeth.2018.05.023
Chemical cross-linking in the structural analysis of protein assemblies
Abstract
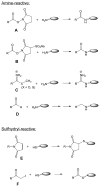
For decades, chemical cross-linking of proteins has been an established method to study protein interaction partners. The chemical cross-linking approach has recently been revived by mass spectrometric analysis of the cross-linking reaction products. Chemical cross-linking and mass spectrometric analysis (CXMS) enables the identification of residues that are close in three-dimensional (3D) space but not necessarily close in primary sequence. Therefore, this approach provides medium resolution information to guide de novo structure prediction, protein interface mapping and protein complex model building. The robustness and compatibility of the CXMS approach with multiple biochemical methods have made it especially appealing for challenging systems with multiple biochemical compositions and conformation states. This review provides an overview of the CXMS approach, describing general procedures in sample processing, data acquisition and analysis. Selection of proper chemical cross-linking reagents, strategies for cross-linked peptide identification, and successful application of CXMS in structural characterization of proteins and protein complexes are discussed.
Keywords: Chemical cross-linking; Integrative modeling; Mass spectrometry.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Advances in protein complex analysis by chemical cross-linking coupled with mass spectrometry (CXMS) and bioinformatics.Biochim Biophys Acta. 2016 Jan;1864(1):123-9. doi: 10.1016/j.bbapap.2015.05.015. Epub 2015 May 27. Biochim Biophys Acta. 2016. PMID: 26025770 Review.
-
Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes.J Mass Spectrom. 2003 Dec;38(12):1225-37. doi: 10.1002/jms.559. J Mass Spectrom. 2003. PMID: 14696200
-
Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique.Anal Chem. 2010 Apr 1;82(7):2636-42. doi: 10.1021/ac1000724. Anal Chem. 2010. PMID: 20210330
-
Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions.Mass Spectrom Rev. 2006 Jul-Aug;25(4):663-82. doi: 10.1002/mas.20082. Mass Spectrom Rev. 2006. PMID: 16477643 Review.
-
Carboxylate-Selective Chemical Cross-Linkers for Mass Spectrometric Analysis of Protein Structures.Anal Chem. 2018 Jan 16;90(2):1195-1201. doi: 10.1021/acs.analchem.7b03789. Epub 2018 Jan 4. Anal Chem. 2018. PMID: 29251911
Cited by
-
Understanding the invisible hands of sample preparation for cryo-EM.Nat Methods. 2021 May;18(5):463-471. doi: 10.1038/s41592-021-01130-6. Epub 2021 May 7. Nat Methods. 2021. PMID: 33963356 Review.
-
Rationally designed bioactive milk-derived protein scaffolds enhanced new bone formation.Bioact Mater. 2022 Jun 16;20:368-380. doi: 10.1016/j.bioactmat.2022.05.028. eCollection 2023 Feb. Bioact Mater. 2022. PMID: 35784638 Free PMC article.
-
A Tailored Strategy to Crosslink the Aspartate Transcarbamoylase Domain of the Multienzymatic Protein CAD.Molecules. 2023 Jan 9;28(2):660. doi: 10.3390/molecules28020660. Molecules. 2023. PMID: 36677714 Free PMC article.
-
Role of integrative structural biology in understanding transcriptional initiation.Methods. 2019 Apr 15;159-160:4-22. doi: 10.1016/j.ymeth.2019.03.009. Epub 2019 Mar 16. Methods. 2019. PMID: 30890443 Free PMC article. Review.
-
Chemical cross-linking and mass spectrometry enabled systems-level structural biology.Curr Opin Struct Biol. 2024 Aug;87:102872. doi: 10.1016/j.sbi.2024.102872. Epub 2024 Jun 26. Curr Opin Struct Biol. 2024. PMID: 38936319 Free PMC article. Review.
References
-
- Busenlehner LS, Armstrong RN. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch Biochem Biophys. 2005;433(1):34–46. - PubMed
-
- Karplus M. Dynamics of proteins. Adv Biophys. 1984;18:165–90. - PubMed
-
- Lee AY, Gulnik SV, Erickson JW. Conformational switching in an aspartic proteinase. Nat Struct Biol. 1998;5(10):866–71. - PubMed
-
- Vetter IR, Wittinghofer A. Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q Rev Biophys. 1999;32(1):1–56. - PubMed
-
- Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991;218(1):233–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

