Regulation of Cytosolic Sulfotransferases in Models of Human Hepatocyte Development
- PMID: 29858374
- PMCID: PMC6038032
- DOI: 10.1124/dmd.118.081398
Regulation of Cytosolic Sulfotransferases in Models of Human Hepatocyte Development
Abstract
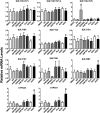
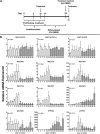
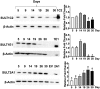
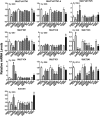
Cytosolic sulfotransferases (SULTs) are expressed during early life and therefore metabolize endogenous and xenobiotic chemicals during development. Little is currently known about the regulation of individual SULTs in the developing human liver. We characterized SULT expression in primary cultures of human fetal hepatocytes and the HepaRG model of liver cell differentiation. SULT1A1 (transcript variants 1-4), SULT1C2, SULT1C4, SULT1E1, and SULT2A1 were the most abundant transcripts in human fetal hepatocytes. In HepaRG cells, SULT1B1, SULT1C2/3/4, and SULT1E1 mRNA levels increased during the transition from proliferation to confluency and then decreased as the cells underwent further differentiation. By contrast, SULT2A1 mRNA levels increased during differentiation, whereas SULT1A1 and SULT2B1 mRNA levels remained relatively constant. The temporal patterns of SULT1C2, SULT1E1, and SULT2A1 protein content were consistent with those observed at the mRNA level. To identify regulators of SULT expression, cultured fetal hepatocytes and HepaRG cells were treated with a panel of lipid- and xenobiotic-sensing receptor activators. The following effects were observed in both fetal hepatocytes and HepaRG cells: 1) liver X receptor activator treatment increased SULT1A1 transcript variant 5 levels; 2) vitamin D receptor activator treatment increased SULT1C2 and SULT2B1 mRNA levels; and 3) farnesoid X receptor activator treatment decreased SULT2A1 expression. Activators of aryl hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, and peroxisome proliferator-activated receptors produced additional gene-dependent effects on SULT expression in HepaRG cells. These findings suggest that SULT-regulating chemicals have the potential to modulate physiologic processes and susceptibility to xenobiotic stressors in the developing human liver.
Copyright © 2018 by The American Society for Pharmacology and Experimental Therapeutics.
Figures


References
-
- Aninat C, Piton A, Glaise D, Le Charpentier T, Langouët S, Morel F, Guguen-Guillouzo C, Guillouzo A. (2006) Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34:75–83. - PubMed
-
- Barker EV, Hume R, Hallas A, Coughtrie WH. (1994) Dehydroepiandrosterone sulfotransferase in the developing human fetus: quantitative biochemical and immunological characterization of the hepatic, renal, and adrenal enzymes. Endocrinology 134:982–989. - PubMed
-
- Berger U, Wilson P, McClelland RA, Colston K, Haussler MR, Pike JW, Coombes RC. (1988) Immunocytochemical detection of 1,25-dihydroxyvitamin D receptors in normal human tissues. J Clin Endocrinol Metab 67:607–613. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

