Structural insights into Rhino-Deadlock complex for germline piRNA cluster specification
- PMID: 29858487
- PMCID: PMC6030695
- DOI: 10.15252/embr.201745418
Structural insights into Rhino-Deadlock complex for germline piRNA cluster specification
Abstract
PIWI-interacting RNAs (piRNAs) silence transposons in germ cells to maintain genome stability and animal fertility. Rhino, a rapidly evolving heterochromatin protein 1 (HP1) family protein, binds Deadlock in a species-specific manner and so defines the piRNA-producing loci in the Drosophila genome. Here, we determine the crystal structures of Rhino-Deadlock complex in Drosophila melanogaster and simulans In both species, one Rhino binds the N-terminal helix-hairpin-helix motif of one Deadlock protein through a novel interface formed by the beta-sheet in the Rhino chromoshadow domain. Disrupting the interface leads to infertility and transposon hyperactivation in flies. Our structural and functional experiments indicate that electrostatic repulsion at the interaction interface causes cross-species incompatibility between the sibling species. By determining the molecular architecture of this piRNA-producing machinery, we discover a novel HP1-partner interacting mode that is crucial to piRNA biogenesis and transposon silencing. We thus explain the cross-species incompatibility of two sibling species at the molecular level.
Keywords: Deadlock; HP1; Rhino; chromoshadow domain; cross‐species incompatibility; piRNA cluster.
© 2018 The Authors.
Figures

- A, B
Top: Domain architecture of Drosophila melanogaster Deadlock (Dm‐Del) and D. simulans Deadlock (Ds‐Del). The N‐terminal 60 amino acids are colored in pink and teal, respectively. Bottom: Schematic of D. melanogaster Rhino (Dm‐Rhi) and D. simulans Rhino (Ds‐Rhi). The chromodomain (CD) is colored in yellow. Dm‐Rhi‐CSD (residue 353–418) is colored in green, and Ds‐Rhi‐CSD (residue 436–500) is colored in violet. Dashed lines indicate the interaction region between Rhi and Del.
- C
The overall structure of the Dm‐Rhi‐CSD in complex with Dm‐Del‐N shown in a ribbon representation and view of 90° rotation around horizontal axis. Dm‐Rhi‐CSD (A: green, B: light blue) form a homodimer. Two molecules of Dm‐Del‐N (A: pink, B: yellow) bind to each monomer of a Dm‐Rhi‐CSD dimer.
- D
Surface representation of Dm‐Rhi‐CSD in complex with Dm‐Del‐N. Color is shown as (C).
- E
Interaction details of Dm‐Rhi‐CSD dimerization.
- F
Dm‐Del‐N represents a helix–hairpin–helix fold. Schematic diagram showing the hydrophobic interaction details between α1 and α2 of Dm‐Del‐N.

- A–D
Yeast two‐hybrid assays mapped the interaction regions between Rhi and Del. Interactions were determined by measuring the β‐galactosidase activity produced by the reporter gene. Data are averages of three independent β‐galactosidase measurements (n = 3) : − (0.00–0.99), + (1.00–9.99), ++ (10.00–99.99), and +++ (100.00–999.99). (A) Dm‐Rhi‐CSD (residues 353–418, colored in green) is sufficient to interact with the N‐terminal 660 amino acids of Del (Dm‐Del‐ΔC). (B) The N‐terminal 60 amino acids of Del (Dm‐Del‐N, colored in pink) is sufficient to interact with Dm‐Rhi‐CSD. (C) Ds‐Rhi‐CSD (residues 436–500, colored in wine) showed no interaction with Dm‐Del‐N. (D) Both Ds‐Rhi‐CSD and Dm‐Rhi‐CSD could interact with Ds‐Del‐N (residues 1–60, colored in teal).
- E
His‐Dm‐Rhi‐CSD and GST‐Dm‐Del‐N form a complex in solution. Left: His‐Dm‐Rhi‐CSD and GST‐Dm‐Del‐N co‐migrated as a single peak on size‐exclusive chromatography (Superdex 75 increase 10/300 column) earlier than His‐Dm‐Rhi‐CSD alone. The elution profiles are indicated in the key.
- F
Top and bottom panels showed the SDS–PAGE results of the peak fractions as indicated in (E).
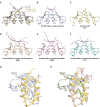
- A
Superimposition of Dm‐Rhi‐CSD (light blue), Dm‐HP1a‐CSD (3P7J, yellow), and Hs‐HP1β‐CSD (3Q6S, wheat).
- B–F
Homodimer formed by CSD domain of HP1 family proteins. Structures are shown with ribbon cartoon. (B) Dm‐Rhi‐CSD in complex with Del. Hydrophobic interface I has not been shown. (C) Dm‐HP1a‐CSD: 3P7J. (D) Hs‐HP1β‐CSD in complex with Shugoshin 1: 3Q6S. (E) Hs‐HP1β‐CSD in complex with EMSY: 2FMM. (F) Hs‐HP1γ‐CSD in complex with histone H3: 5T1I.
- G
Detailed intermolecular contacts on interface I’ formed by Dm‐Del‐N (B: yellow) and Dm‐Rhi‐CSD (B: light blue).
- H
Superimposition of interface I and I’.
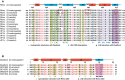
Sequence alignment of CSDs in Rhino and other HP1 family members. Amino acids that are identical and conserved are shown in red and pink. Residues on the Rhi‐CSD dimeric interface are indicated by blue squares. Residues involved in interface I and interface II are labeled by green triangles and purple pentagons, respectively. The amino acid difference causing conformational change between Dm‐Rhi‐CSD and Ds‐Rhi‐CSD is framed in red. Secondary structural elements of Dm‐Rhi‐CSD are indicated above.
Sequence alignment of Del‐N in Drosophila species. Color code is indicated as in (A). Residues forming the interface I and interface II are marked by green triangles and purple pentagons, respectively. Secondary structural elements of Dm‐Del‐N are indicated above.

- A
Interaction interfaces between Dm‐Del‐N (A: pink), Dm‐Del‐N (B: yellow), and Dm‐Rhi‐CSD homodimer (A: green, B: light blue). Interface I is formed by the HhH motif of Dm‐Del‐N (A) and the β‐sheets of Dm‐Rhi‐CSD (A). Interface I’ is formed by Dm‐Del‐N (B) and Dm‐Rhi‐CSD (B). Interface II consists of the β‐sandwich structure formed by the C‐terminal extension of Dm‐Del‐N (A) and the C‐terminal β‐strands of Dm‐Rhi‐CSD dimer.
- B, C
Stereo views of detailed intermolecular contacts on interface I and interface II, respectively. Yellow dashed lines indicate the hydrogen bonds.
- D
A surface representation showing the C‐terminal β‐strand of Dm‐Del‐N (A: pink) is located at the center of Dm‐Rhi‐CSD dimer. I50 occupies the central position and is named as position 0, Y48, C49, Q51, and T52 are designated as position −2, −1, +1, and +2. Dm‐Del‐N (A) and Dm‐Del‐N (B) are shown in a cartoon model.
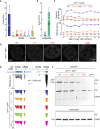
Yeast two‐hybrid assays indicate the binding of wild‐type or mutant Dm‐Rhi‐CSD with Dm‐Del‐N. Mutations of key residues on both interface I or interface II abolish the binding. The averages of three independent β‐galactosidase measurements (n = 3) with SD values (error bar) are shown. The detailed β‐gal units are listed as follows: WT, 7.95 ± 3.10; YFF, 0.28 ± 0.02; HYM, 0.61 ± 0.03; LRIIV, 0.60 ± 0.09; NDD, 0.24 ± 0.07; HIND, 0.44 ± 0.37; K28M, 1.85 ± 0.20.
Reverse mutant of Ds‐Rhi‐CSD could interact with Dm‐Del‐N. The averages of three independent β‐galactosidase measurements (n = 3) with SD values (error bar) are shown. The detailed β‐gal units are listed as follows: WT, 0.40 ± 0.04; RM, 60.50 ± 4.64.
Fertility of the rhi mutant flies carrying one copy of transgene expresses either wild‐type Rhi or Rhi with amino acid mutations. The transgene is driven by rhi promoter and has an eGFP tag at N‐terminus.
The localization of wild‐type Rhi or Rhi with amino acid mutations.
RNA‐Seq to quantify the expression of Rhi transcripts from each transgene. RhoGAP54D and CG30105 are two genes locate upstream of rhi. The transcripts from them serve as normalization control. rhi 2/KG flies only produce a few truncated rhi transcripts. Note: The GFP tagged rhi construct does not contain 3'UTR of rhi.
Western blot to quantify the GFP::Rhi proteins from 60 fly ovaries.
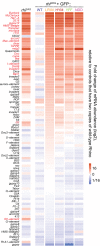

Stereo views of the hydrophobic interaction details between two α‐helices of Ds‐Del‐NΔβ. Residues involved in the hydrophobic interaction are labeled. I7, M9, L13, W16, I19, L20, L23 on α1 and M28, W33, F36, F37, F40, L41, W44 on α2 form a hydrophobic cluster. W44 and K12 form a cation‐π interaction.
Superimposition of Ds‐Del‐NΔβ (lime) and Dm‐Del‐N (pink).
Stereo views of the interface I between Ds‐Rhi‐CSDΔβ (wine) and Ds‐Del‐NΔβ (lime). Residues involved in the interface are labeled. F40, W44 on Ds‐Del‐NΔβ, and H452, Y454, F460, F462 on Ds‐Rhi‐CSDΔβ form an aromatic cluster. C15 on Ds‐Del‐NΔβ and A473 on Ds‐Rhi‐CSDΔβ further enhance the aromatic cluster. I7 on Ds‐Del‐NΔβ has hydrophobic interactions with L451 and I471 on Ds‐Rhi‐CSDΔβ. M456 on Ds‐Rhi‐CSDΔβ is surrounded by I19, L23, F36 on Ds‐Del‐NΔβ.
Superimposition of interface I between Rhi‐CSD and Del‐N in Drosophila melanogaster and Drosophila simulans.
The 2Fo‐Fc electron density map (contoured at 1.0 σ Cutoff, blue mesh) of residues involved in the interactions between L1 of Rhi‐CSD and η1 of Del‐N in Drosophila simulans (left) and Drosophila melanogaster (right).

Overall structure of the Ds‐Rhi‐CSDΔβ in complex with Ds‐Del‐NΔβ shown in a ribbon representation. Ds‐Rhi‐CSDΔβ is colored in wine, and Ds‐Del‐N is colored in lime.
SEC‐MALS assay of the covalently linked Rhi‐CSD and Del‐N fusion proteins as indicated in the key.
Superposition of melanogaster and simulans Rhi‐CSD and Del‐N complexes. Colors are indicated in the key. The L1 and L2 loops are circled in black.
Residues on L2 interact with L1 and stabilize L1 both in simulans (left) and in melanogaster (right).
Details of the intermolecular interactions between L1 of Rhi‐CSD and η1 of Del‐N. K457 on Ds‐Rhi‐CSDΔβ forms a hydrogen bond with S27 on Ds‐Del‐NΔβ (left). D375 and D376 on Dm‐Rhi‐CSD form electrostatic interactions with K28 on Dm‐Del‐N (right).
Left: Ds‐Rhi‐CSDΔβ binds to Ds‐Del‐NΔβ. Right: Dm‐Rhi‐CSD binds to Dm‐Del‐N. The CSDs are shown in cartoon representation. The L1 site on each structure is shown in stick mode. Ds‐Del‐N has a more negatively charged surface area within the L1 binding site than that in Dm‐Del‐N (electrostatic potential level is set from −50 kT/e to +50 kT/e).
References
-
- Weick EM, Miska EA (2014) piRNAs: from biogenesis to function. Development 141: 3458–3471 - PubMed
-
- Siomi MC, Sato K, Pezic D, Aravin AA (2011) PIWI‐interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 - PubMed
-
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

