The structure of human Nocturnin reveals a conserved ribonuclease domain that represses target transcript translation and abundance in cells
- PMID: 29860338
- PMCID: PMC6158716
- DOI: 10.1093/nar/gky412
The structure of human Nocturnin reveals a conserved ribonuclease domain that represses target transcript translation and abundance in cells
Abstract
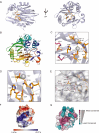
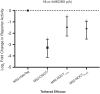
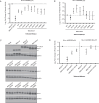
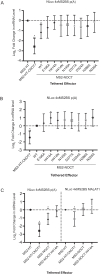
The circadian protein Nocturnin (NOCT) belongs to the exonuclease, endonuclease and phosphatase superfamily and is most similar to the CCR4-class of deadenylases that degrade the poly-adenosine tails of mRNAs. NOCT-deficient mice are resistant to high-fat diet induced weight gain, and exhibit dysregulation of bone formation. However, the mechanisms by which NOCT regulates these processes remain to be determined. Here, we describe a pair of high-resolution crystal structures of the human NOCT catalytic domain. The active site of NOCT is highly conserved with other exoribonucleases, and when directed to a transcript in cells, NOCT can reduce translation and abundance of that mRNA in a manner dependent on key active site residues. In contrast to the related deadenylase CNOT6L, purified recombinant NOCT lacks in vitro ribonuclease activity, suggesting that unidentified factors are necessary for enzymatic activity. We also find the ability of NOCT to repress reporter mRNAs in cells depends upon the 3' end of the mRNA, as reporters terminating with a 3' MALAT1 structure cannot be repressed by NOCT. Together, these data demonstrate that NOCT is an exoribonuclease that can degrade mRNAs to inhibit protein expression, suggesting a molecular mechanism for its regulatory role in lipid metabolism and bone development.
Figures

Similar articles
-
Crystal Structure of Human Nocturnin Catalytic Domain.Sci Rep. 2018 Nov 2;8(1):16294. doi: 10.1038/s41598-018-34615-0. Sci Rep. 2018. PMID: 30389976 Free PMC article.
-
Differential processing and localization of human Nocturnin controls metabolism of mRNA and nicotinamide adenine dinucleotide cofactors.J Biol Chem. 2020 Oct 30;295(44):15112-15133. doi: 10.1074/jbc.RA120.012618. Epub 2020 Aug 23. J Biol Chem. 2020. PMID: 32839274 Free PMC article.
-
A novel mouse model overexpressing Nocturnin results in decreased fat mass in male mice.J Cell Physiol. 2019 Nov;234(11):20228-20239. doi: 10.1002/jcp.28623. Epub 2019 Apr 5. J Cell Physiol. 2019. PMID: 30953371 Free PMC article.
-
Circadian control of Nocturnin and its regulatory role in health and disease.Chronobiol Int. 2023 Jul 3;40(7):970-981. doi: 10.1080/07420528.2023.2231081. Epub 2023 Jul 3. Chronobiol Int. 2023. PMID: 37400970 Review.
-
Kiss your tail goodbye: the role of PARN, Nocturnin, and Angel deadenylases in mRNA biology.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):571-9. doi: 10.1016/j.bbagrm.2012.12.004. Epub 2012 Dec 26. Biochim Biophys Acta. 2013. PMID: 23274303 Free PMC article. Review.
Cited by
-
The metabolites NADP+ and NADPH are the targets of the circadian protein Nocturnin (Curled).Nat Commun. 2019 May 30;10(1):2367. doi: 10.1038/s41467-019-10125-z. Nat Commun. 2019. PMID: 31147539 Free PMC article.
-
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.Biochemistry. 2022 May 10:10.1021/acs.biochem.2c00072. doi: 10.1021/acs.biochem.2c00072. Online ahead of print. Biochemistry. 2022. PMID: 35535990 Free PMC article.
-
Human Pumilio proteins directly bind the CCR4-NOT deadenylase complex to regulate the transcriptome.RNA. 2021 Apr;27(4):445-464. doi: 10.1261/rna.078436.120. Epub 2021 Jan 4. RNA. 2021. PMID: 33397688 Free PMC article.
-
Regulatory roles of vertebrate Nocturnin: insights and remaining mysteries.RNA Biol. 2018;15(10):1255-1267. doi: 10.1080/15476286.2018.1526541. Epub 2018 Oct 9. RNA Biol. 2018. PMID: 30257600 Free PMC article.
-
Crystal Structure of Human Nocturnin Catalytic Domain.Sci Rep. 2018 Nov 2;8(1):16294. doi: 10.1038/s41598-018-34615-0. Sci Rep. 2018. PMID: 30389976 Free PMC article.
References
-
- Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007; 8:113–126. - PubMed
-
- Goldstrohm A.C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008; 9:337–344. - PubMed
-
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006; 127:1223–1237. - PubMed
-
- Geisler S., Coller J. XRN1: a Major 5′ to 3′ exoribonuclease in eukaryotic cells. Enzymes. 2012; 31:97–114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

